Genome rearrangements and selection in multi-chromosome bacteria Burkholderia spp
- PMID: 30587126
- PMCID: PMC6307245
- DOI: 10.1186/s12864-018-5245-1
Genome rearrangements and selection in multi-chromosome bacteria Burkholderia spp
Abstract
Background: The genus Burkholderia consists of species that occupy remarkably diverse ecological niches. Its best known members are important pathogens, B. mallei and B. pseudomallei, which cause glanders and melioidosis, respectively. Burkholderia genomes are unusual due to their multichromosomal organization, generally comprised of 2-3 chromosomes.
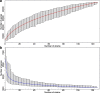
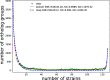
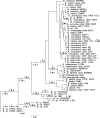
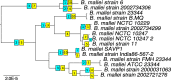
Results: We performed integrated genomic analysis of 127 Burkholderia strains. The pan-genome is open with the saturation to be reached between 86,000 and 88,000 genes. The reconstructed rearrangements indicate a strong avoidance of intra-replichore inversions that is likely caused by selection against the transfer of large groups of genes between the leading and the lagging strands. Translocated genes also tend to retain their position in the leading or the lagging strand, and this selection is stronger for large syntenies. Integrated reconstruction of chromosome rearrangements in the context of strains phylogeny reveals parallel rearrangements that may indicate inversion-based phase variation and integration of new genomic islands. In particular, we detected parallel inversions in the second chromosomes of B. pseudomallei with breakpoints formed by genes encoding membrane components of multidrug resistance complex, that may be linked to a phase variation mechanism. Two genomic islands, spreading horizontally between chromosomes, were detected in the B. cepacia group.
Conclusions: This study demonstrates the power of integrated analysis of pan-genomes, chromosome rearrangements, and selection regimes. Non-random inversion patterns indicate selective pressure, inversions are particularly frequent in a recent pathogen B. mallei, and, together with periods of positive selection at other branches, may indicate adaptation to new niches. One such adaptation could be a possible phase variation mechanism in B. pseudomallei.
Keywords: Burkholderia; Comparative genomics; Genome rearrangements; Multi-chromosome bacteria; Pan-genome; Positive selection; Strain phylogeny.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

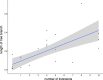



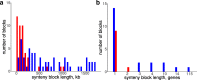
References
-
- Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5:719–29. - PubMed
-
- Howe C, Sampath A, Spotnitz M. The pseudomallei group: a review. J Infect Dis. 1971;124:598–606. - PubMed
-
- Goris J, De Vos P, Caballero-Mellado J, Park J, Falsen E, Quensen Jr, Tiedje J, Vandamme P. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int J Syst Evol Microbiol. 2004;54:1677–81. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

