Detection of viable but non-culturable Pseudomonas aeruginosa in cystic fibrosis by qPCR: a validation study
- PMID: 30587160
- PMCID: PMC6307279
- DOI: 10.1186/s12879-018-3612-9
Detection of viable but non-culturable Pseudomonas aeruginosa in cystic fibrosis by qPCR: a validation study
Abstract
Background: Routine culture-based diagnosis of Pseudomonas aeruginosa lung infection in Cystic Fibrosis (CF) patients can be hampered by the phenotypic variability of the microorganism, including its transition to a Viable But Non-Culturable (VBNC) state. The aim of this study was to validate an ecfX-targeting qPCR protocol developed to detect all viable P. aeruginosa bacteria and to identify VBNC forms in CF sputum samples.
Methods: The study involved 115 P. aeruginosa strains of different origins and 10 non-P. aeruginosa strains and 88 CF sputum samples, 41 Culture-Positive (CP) and 47 Culture-Negative (CN). Spiking assays were performed using scalar dilutions of a mixture of live and dead P. aeruginosa ATCC 9027 and a pooled P. aeruginosa-free sputum batch. Total DNA from sputum samples was extracted by a commercial kit, whereas a crude extract was obtained from the broth cultures. Extracellular DNA (eDNA) interference was evaluated by comparing the qPCR counts obtained from DNase-treated and untreated aliquots of the same samples. The statistical significance of the results was assessed by the Wilcoxon test and Student's t test.
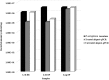
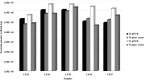
Results: The newly-developed qPCR protocol identified 96.6% of the P. aeruginosa isolates; no amplification was obtained with strains belonging to different species. Spiking assays supported protocol reliability, since counts always matched the amount of live bacteria, thus excluding the interference of dead cells and eDNA. The protocol sensitivity threshold was 70 cells/ml of the original sample. Moreover, qPCR detected P. aeruginosa in 9/47 CN samples and showed higher bacterial counts compared with the culture method in 10/41 CP samples.
Conclusions: Our findings demonstrate the reliability of the newly-developed qPCR protocol and further highlight the need for harnessing a non-culture approach to achieve an accurate microbiological diagnosis of P. aeruginosa CF lung infection and a greater understanding of its evolution.
Keywords: Cystic fibrosis; Lung infection; Pseudomonas aeruginosa; Viable but non-culturable bacterial forms; qPCR.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Greipel L, Fischer S, Klockgether J, Dorda M, Mielke S, Wiehlmann L, et al. Molecular epidemiology of mutations in antimicrobial resistance loci of Pseudomonas aeruginosa isolates from airways of cystic fibrosis patients. Antimicrob Agents Chemother. 2016;60(11):6726–6734. doi: 10.1128/AAC.00724-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

