Bipartite promoters and RNA editing of paramyxoviruses and filoviruses
- PMID: 30587495
- PMCID: PMC6380270
- DOI: 10.1261/rna.068825.118
Bipartite promoters and RNA editing of paramyxoviruses and filoviruses
Abstract
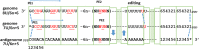
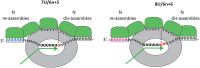
A primary property of paramyxovirus bipartite promoters is to ensure that their RNA genomes are imprinted with a hexamer phase via their association with nucleoproteins, in part because this phase as well the editing sequence itself controls mRNA editing. The question then arises whether a similar mechanism operates for filoviruses that also contain bipartite promoters that are governed by the "rule of six," even though these genomes need not, and given Ebola virus biology, cannot always be of hexamer genome length. This review suggests that this is possible and describes how it might operate, and that RNA editing may play a role in Ebola virus genome interconversion that helps the virus adapt to different host environments.
Keywords: Ebola virus; RNA editing; bipartite promoters; genome interconversion; rule of six.
© 2019 le Mercier and Kolakofsky; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


Similar articles
-
Paramyxovirus RNA synthesis, mRNA editing, and genome hexamer phase: A review.Virology. 2016 Nov;498:94-98. doi: 10.1016/j.virol.2016.08.018. Epub 2016 Aug 25. Virology. 2016. PMID: 27567257 Review.
-
Phylogenetic analysis of the promoter element 2 of paramyxo- and filoviruses.Microbiol Spectr. 2024 May 2;12(5):e0041724. doi: 10.1128/spectrum.00417-24. Epub 2024 Apr 12. Microbiol Spectr. 2024. PMID: 38606982 Free PMC article.
-
The control of paramyxovirus genome hexamer length and mRNA editing.RNA. 2018 Apr;24(4):461-467. doi: 10.1261/rna.065243.117. Epub 2018 Jan 22. RNA. 2018. PMID: 29358233 Free PMC article.
-
Paramyxovirus RNA editing and the requirement for hexamer genome length.RNA. 1996 Oct;2(10):1033-45. RNA. 1996. PMID: 8849779 Free PMC article.
-
Molecular biology and evolution of filoviruses.Arch Virol Suppl. 1993;7:81-100. doi: 10.1007/978-3-7091-9300-6_8. Arch Virol Suppl. 1993. PMID: 8219816 Review.
Cited by
-
Identification and characterization of short leader and trailer RNAs synthesized by the Ebola virus RNA polymerase.PLoS Pathog. 2021 Oct 26;17(10):e1010002. doi: 10.1371/journal.ppat.1010002. eCollection 2021 Oct. PLoS Pathog. 2021. PMID: 34699554 Free PMC article.
-
Negative and ambisense RNA virus ribonucleocapsids: more than protective armor.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0008223. doi: 10.1128/mmbr.00082-23. Epub 2023 Sep 26. Microbiol Mol Biol Rev. 2023. PMID: 37750733 Free PMC article. Review.
-
A Comparative Assessment of the Pathogenic Potential of Newly Discovered Henipaviruses.Pathogens. 2024 Jul 16;13(7):587. doi: 10.3390/pathogens13070587. Pathogens. 2024. PMID: 39057814 Free PMC article. Review.
-
Evidence that hPIV2 paramyxovirus antigenomes are edited during infection.mBio. 2025 Aug 13;16(8):e0366724. doi: 10.1128/mbio.03667-24. Epub 2025 Jun 24. mBio. 2025. PMID: 40552829 Free PMC article.
-
Phylodynamic study of the conserved RNA structure encompassing the hemagglutinin cleavage site encoding region of H5 and H7 low pathogenic avian influenza viruses.Virus Evol. 2021 Nov 1;7(2):veab093. doi: 10.1093/ve/veab093. eCollection 2021 Sep. Virus Evol. 2021. PMID: 35299790 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
