Pathogenicity and selective constraint on variation near splice sites
- PMID: 30587507
- PMCID: PMC6360807
- DOI: 10.1101/gr.238444.118
Pathogenicity and selective constraint on variation near splice sites
Abstract
Mutations that perturb normal pre-mRNA splicing are significant contributors to human disease. We used exome sequencing data from 7833 probands with developmental disorders (DDs) and their unaffected parents, as well as more than 60,000 aggregated exomes from the Exome Aggregation Consortium, to investigate selection around the splice sites and quantify the contribution of splicing mutations to DDs. Patterns of purifying selection, a deficit of variants in highly constrained genes in healthy subjects, and excess de novo mutations in patients highlighted particular positions within and around the consensus splice site of greater functional relevance. By using mutational burden analyses in this large cohort of proband-parent trios, we could estimate in an unbiased manner the relative contributions of mutations at canonical dinucleotides (73%) and flanking noncanonical positions (27%), and calculate the positive predictive value of pathogenicity for different classes of mutations. We identified 18 patients with likely diagnostic de novo mutations in dominant DD-associated genes at noncanonical positions in splice sites. We estimate 35%-40% of pathogenic variants in noncanonical splice site positions are missing from public databases.
© 2019 Lord et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

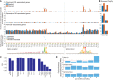
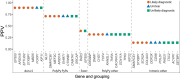
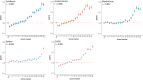

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
