Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis
- PMID: 30587782
- PMCID: PMC6337374
- DOI: 10.3390/ijms20010082
Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis
Abstract
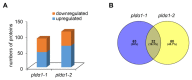
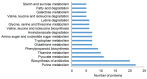
Phospholipase Dα1 (PLDα1) belongs to phospholipases, a large phospholipid hydrolyzing protein family. PLDα1 has a substrate preference for phosphatidylcholine leading to enzymatic production of phosphatidic acid, a lipid second messenger with multiple cellular functions. PLDα1 itself is implicated in biotic and abiotic stress responses. Here, we present a shot-gun differential proteomic analysis on roots of two Arabidopsis pldα1 mutants compared to the wild type. Interestingly, PLDα1 deficiency leads to altered abundances of proteins involved in diverse processes related to membrane transport including endocytosis and endoplasmic reticulum-Golgi transport. PLDα1 may be involved in the stability of attachment sites of endoplasmic reticulum to the plasma membrane as suggested by increased abundance of synaptotagmin 1, which was validated by immunoblotting and whole-mount immunolabelling analyses. Moreover, we noticed a robust abundance alterations of proteins involved in mitochondrial import and electron transport chain. Notably, the abundances of numerous proteins implicated in glucosinolate biosynthesis were also affected in pldα1 mutants. Our results suggest a broader biological involvement of PLDα1 than anticipated thus far, especially in the processes such as endomembrane transport, mitochondrial protein import and protein quality control, as well as glucosinolate biosynthesis.
Keywords: Arabidopsis; cytoskeleton; mitochondrial protein import; phospholipase D alpha1; proteomics; quality control; vesicular transport.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Wang X., editor. Phospholipases in Plant Signaling. Volume 20. Springer; Berlin/Heidelberg, Germany: 2014. (Signaling and Communication in Plants).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

