The Unique Mechanisms of Cellular Proliferation, Migration and Apoptosis are Regulated through Oocyte Maturational Development-A Complete Transcriptomic and Histochemical Study
- PMID: 30587792
- PMCID: PMC6337548
- DOI: 10.3390/ijms20010084
The Unique Mechanisms of Cellular Proliferation, Migration and Apoptosis are Regulated through Oocyte Maturational Development-A Complete Transcriptomic and Histochemical Study
Abstract
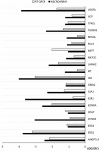
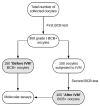
The growth and development of oocyte affect the functional activities of the surrounding somatic cells. These cells are regulated by various types of hormones, proteins, metabolites, and regulatory molecules through gap communication, ultimately leading to the development and maturation of oocytes. The close association between somatic cells and oocytes, which together form the cumulus-oocyte complexes (COCs), and their bi-directional communication are crucial for the acquisition of developmental competences by the oocyte. In this study, oocytes were extracted from the ovaries obtained from crossbred landrace gilts and subjected to in vitro maturation. RNA isolated from those oocytes was used for the subsequent microarray analysis. The data obtained shows, for the first time, variable levels of gene expression (fold changes higher than |2| and adjusted p-value < 0.05) belonging to four ontological groups: regulation of cell proliferation (GO:0042127), regulation of cell migration (GO:0030334), and regulation of programmed cell death (GO:0043067) that can be used together as proliferation, migration or apoptosis markers. We have identified several genes of porcine oocytes (ID2, VEGFA, BTG2, ESR1, CCND2, EDNRA, ANGPTL4, TGFBR3, GJA1, LAMA2, KIT, TPM1, VCP, GRID2, MEF2C, RPS3A, PLD1, BTG3, CD47, MITF), whose expression after in vitro maturation (IVM) is downregulated with different degrees. Our results may be helpful in further elucidating the molecular basis and functional significance of a number of gene markers associated with the processes of migration, proliferation and angiogenesis occurring in COCs.
Keywords: cellular competence; microarray; oocytes; pig.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
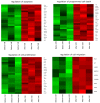

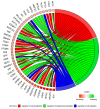


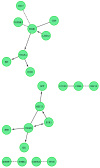
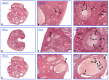
References
-
- Sinha P.B., Tesfaye D., Rings F., Hossien M., Hoelker M., Held E., Neuhoff C., Tholen E., Schellander K., Salilew-Wondim D. MicroRNA-130b is involved in bovine granulosa and cumulus cells function, oocyte maturation and blastocyst formation. J. Ovarian Res. 2017;10:37. doi: 10.1186/s13048-017-0336-1. - DOI - PMC - PubMed
-
- Kempisty B., Ziółkowska A., Ciesiółka S., Piotrowska H., Antosik P., Bukowska D., Nowicki M., Brüssow K.P., Zabel M. Study on connexin gene and protein expression and cellular distribution in relation to real-time proliferation of porcine granulosa cells. J. Biol. Regul. Homeost. Agents. 2014;28:625–635. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

