Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures
- PMID: 30587834
- PMCID: PMC6337575
- DOI: 10.3390/ijms20010092
Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures
Abstract
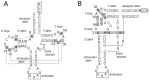
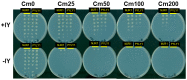
The L-shape form of tRNA is maintained by tertiary interactions occurring in the core. Base changes in this domain can cause structural defects and impair tRNA activity. Here, we report on a method to safely engineer structural variations in this domain utilizing the noncanonical scaffold of tRNAPyl. First, we constructed a naïve hybrid between archaeal tRNAPyl and tRNATyr, which consisted of the acceptor and T stems of tRNATyr and the other parts of tRNAPyl. This hybrid tRNA efficiently translated the UAG codon to 3-iodotyrosine in Escherichia coli cells, when paired with a variant of the archaeal tyrosyl-tRNA synthetase. The amber suppression efficiency was slightly lower than that of the "bench-mark" archaeal tRNATyr suppressor assuming the canonical structure. After a series of modifications to this hybrid tRNA, we obtained two artificial types of tRNATyr: ZtRNA had an augmented D (auD) helix in a noncanonical form and the D and T loops bound by the standard tertiary base pairs, and YtRNA had a canonical auD helix and non-standard interloop interactions. It was then suggested that the ZtRNA scaffold could also support the glycylation and glutaminylation of tRNA. The synthetic diversity of tRNA would help create new tRNA⁻aminoacyl-tRNA synthetase pairs for reprogramming the genetic code.
Keywords: amber suppression; genetic code expansion; pyrrolysine tRNA; tRNA secondary structure; tertiary base pairs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
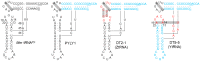
Similar articles
-
tRNAPyl: Structure, function, and applications.RNA Biol. 2018;15(4-5):441-452. doi: 10.1080/15476286.2017.1356561. Epub 2017 Sep 13. RNA Biol. 2018. PMID: 28837402 Free PMC article. Review.
-
Pyrrolysine is not hardwired for cotranslational insertion at UAG codons.Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3141-6. doi: 10.1073/pnas.0611634104. Epub 2007 Feb 20. Proc Natl Acad Sci U S A. 2007. PMID: 17360621 Free PMC article.
-
Evolved sequence contexts for highly efficient amber suppression with noncanonical amino acids.ACS Chem Biol. 2014 Dec 19;9(12):2815-22. doi: 10.1021/cb5006273. Epub 2014 Oct 17. ACS Chem Biol. 2014. PMID: 25299570
-
A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine.Proc Natl Acad Sci U S A. 2007 Jan 16;104(3):1021-6. doi: 10.1073/pnas.0610294104. Epub 2007 Jan 4. Proc Natl Acad Sci U S A. 2007. PMID: 17204561 Free PMC article.
-
Orthogonal Protein Translation Using Pyrrolysyl-tRNA Synthetases for Single- and Multiple-Noncanonical Amino Acid Mutagenesis.Adv Biochem Eng Biotechnol. 2018;162:1-19. doi: 10.1007/10_2016_37. Adv Biochem Eng Biotechnol. 2018. PMID: 27783132 Review.
Cited by
-
Genetic Code Expansion: Another Solution to Codon Assignments.Int J Mol Sci. 2022 Dec 26;24(1):361. doi: 10.3390/ijms24010361. Int J Mol Sci. 2022. PMID: 36613803 Free PMC article.
-
Rational Design of Aptamer-Tagged tRNAs.Int J Mol Sci. 2020 Oct 21;21(20):7793. doi: 10.3390/ijms21207793. Int J Mol Sci. 2020. PMID: 33096801 Free PMC article.
-
Initiating protein synthesis with noncanonical monomers in vitro and in vivo.Methods Enzymol. 2021;656:495-519. doi: 10.1016/bs.mie.2021.05.002. Epub 2021 Jun 2. Methods Enzymol. 2021. PMID: 34325796 Free PMC article.
-
Aminoacyl-tRNA Synthetases and tRNAs for an Expanded Genetic Code: What Makes them Orthogonal?Int J Mol Sci. 2019 Apr 19;20(8):1929. doi: 10.3390/ijms20081929. Int J Mol Sci. 2019. PMID: 31010123 Free PMC article. Review.
-
Structural insights into dynamics of the BMV TLS aminoacylation.Nat Commun. 2025 Feb 3;16(1):1276. doi: 10.1038/s41467-025-56612-4. Nat Commun. 2025. PMID: 39900568 Free PMC article.
References
-
- Dirheimer G., Keith G., Dumas P., Westhof E. Primary, secondary, and tertiary structures of tRNAs. In: Söll D., RajBhandary U.L., editors. tRNA: Structure, Biosynthesis, and Function. AMS Press; Washington, DC, USA: 1995. pp. 93–126.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

