Long non-coding RNA LUCAT1 promotes tumourigenesis by inhibiting ANXA2 phosphorylation in hepatocellular carcinoma
- PMID: 30588744
- PMCID: PMC6378214
- DOI: 10.1111/jcmm.14088
Long non-coding RNA LUCAT1 promotes tumourigenesis by inhibiting ANXA2 phosphorylation in hepatocellular carcinoma
Abstract
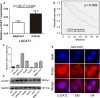
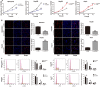
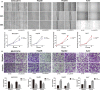
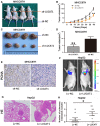
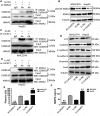
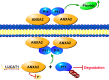
Long non-coding RNAs (lncRNAs) play essential roles in diverse biological processes; however, current understanding of the mechanism underlying the regulation of tumour proliferation and metastasis is limited. Lung cancer-associated transcript 1 (LUCAT1) has been reported in a variety of human cancers, while its role in hepatocellular carcinoma (HCC) remains unclear. This study aimed to determine the biological role and underlying mechanism of LUCAT1 on progression and metastasis in HCC cells and clinical specimens. Our results demonstrated that LUCAT1 was up-regulated in HCC tissues and cells. Loss- and gain-of-function studies revealed that LUCAT1 promotes the proliferation and metastasis of HCC cells in vitro and in vivo. Furthermore, RNA pulldown and Western blot assays indicated that LUCAT1 inhibited the phosphorylation of Annexin A2 (ANXA2) to reduce the degradation of ANXA2-S100A10 heterotetramer (AIIt), which in turn accelerated the secretion of plasminogen into plasmin, thereby resulting in the activation of metalloprotease proteins. In conclusion, we propose that LUCAT1 serves as a novel diagnostic and therapeutic target for HCC.
Keywords: Annexin A2; hepatocellular carcinoma; long non-coding RNA; phosphorylation; tumour progression and metastasis.
© 2018 The Authors Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine.
Figures

Similar articles
-
Long noncoding RNA CCAT2 promotes hepatocellular carcinoma proliferation and metastasis through up-regulation of NDRG1.Exp Cell Res. 2019 Jun 1;379(1):19-29. doi: 10.1016/j.yexcr.2019.03.029. Epub 2019 Mar 25. Exp Cell Res. 2019. PMID: 30922920
-
LINC00174 is an oncogenic lncRNA of hepatocellular carcinoma and regulates miR-320/S100A10 axis.Cell Biochem Funct. 2020 Oct;38(7):859-869. doi: 10.1002/cbf.3498. Epub 2020 Mar 3. Cell Biochem Funct. 2020. PMID: 32128852
-
Long non-coding RNA NRON is downregulated in HCC and suppresses tumour cell proliferation and metastasis.Biomed Pharmacother. 2018 Aug;104:102-109. doi: 10.1016/j.biopha.2018.05.006. Epub 2018 May 15. Biomed Pharmacother. 2018. PMID: 29772429
-
Functional long non-coding RNAs in hepatocellular carcinoma.Cancer Lett. 2021 Mar 1;500:281-291. doi: 10.1016/j.canlet.2020.10.042. Epub 2020 Oct 29. Cancer Lett. 2021. PMID: 33129957 Review.
-
The Role of Long Non-Coding RNA and microRNA Networks in Hepatocellular Carcinoma and Its Tumor Microenvironment.Int J Mol Sci. 2021 Sep 30;22(19):10630. doi: 10.3390/ijms221910630. Int J Mol Sci. 2021. PMID: 34638971 Free PMC article. Review.
Cited by
-
Concomitant and decoupled effects of cigarette smoke and SCAL1 upregulation on oncogenic phenotypes and ROS detoxification in lung adenocarcinoma cells.Sci Rep. 2021 Sep 15;11(1):18345. doi: 10.1038/s41598-021-97869-1. Sci Rep. 2021. PMID: 34526564 Free PMC article.
-
The regulatory role of lncRNA in tumor drug resistance: refracting light through a narrow aperture.Oncol Res. 2025 Mar 19;33(4):837-849. doi: 10.32604/or.2024.053882. eCollection 2025. Oncol Res. 2025. PMID: 40191723 Free PMC article. Review.
-
The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma.Mol Cancer. 2019 Oct 25;18(1):147. doi: 10.1186/s12943-019-1086-z. Mol Cancer. 2019. PMID: 31651347 Free PMC article. Review.
-
Updating the Clinical Application of Blood Biomarkers and Their Algorithms in the Diagnosis and Surveillance of Hepatocellular Carcinoma: A Critical Review.Int J Mol Sci. 2023 Feb 21;24(5):4286. doi: 10.3390/ijms24054286. Int J Mol Sci. 2023. PMID: 36901717 Free PMC article. Review.
-
A novel DNA methylation-driver gene signature for long-term survival prediction of hepatitis-positive hepatocellular carcinoma patients.Cancer Med. 2022 Dec;11(23):4721-4735. doi: 10.1002/cam4.4838. Epub 2022 May 30. Cancer Med. 2022. PMID: 35637633 Free PMC article.
References
-
- Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016;36(3):317‐324. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Takayama T. Surgical treatment for hepatocellular carcinoma. Jpn J Clin Oncol. 2011;41(4):447‐454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

