BioID identifies proteins involved in the cell biology of caveolae
- PMID: 30589899
- PMCID: PMC6307745
- DOI: 10.1371/journal.pone.0209856
BioID identifies proteins involved in the cell biology of caveolae
Abstract
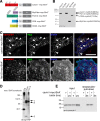
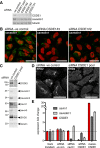
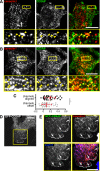
The mechanisms controlling the abundance and sub-cellular distribution of caveolae are not well described. A first step towards determining such mechanisms would be identification of relevant proteins that interact with known components of caveolae. Here, we applied proximity biotinylation (BioID) to identify a list of proteins that may interact with the caveolar protein cavin1. Screening of these candidates using siRNA to reduce their expression revealed that one of them, CSDE1, regulates the levels of mRNAs and protein expression for multiple components of caveolae. A second candidate, CD2AP, co-precipitated with cavin1. Caveolar proteins were observed in characteristic and previously un-described linear arrays adjacent to cell-cell junctions in both MDCK cells, and in HeLa cells overexpressing an active form of the small GTPase Rac1. CD2AP was required for the recruitment of caveolar proteins to these linear arrays. We conclude that BioID will be useful in identification of new proteins involved in the cell biology of caveolae, and that interaction between CD2AP and cavin1 may have an important role in regulating the sub-cellular distribution of caveolae.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

