, Jost's D, and FST are similarly constrained by allele frequencies: A mathematical, simulation, and empirical study
- PMID: 30589985
- PMCID: PMC6821915
- DOI: 10.1111/mec.15000
, Jost's D, and FST are similarly constrained by allele frequencies: A mathematical, simulation, and empirical study
Abstract
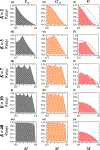
Statistics and Jost's D have been proposed for replacing FST as measures of genetic differentiation. A principal argument in favour of these statistics is the independence of their maximal values with respect to the subpopulation heterozygosity HS , a property not shared by FST . Nevertheless, it has been unclear if these alternative differentiation measures are constrained by other aspects of the allele frequencies. Here, for biallelic markers, we study the mathematical properties of the maximal values of and D, comparing them to those of FST . We show that and D exhibit the same peculiar frequency-dependence phenomena as FST , including a maximal value as a function of the frequency of the most frequent allele that lies well below one. Although the functions describing , D, and FST in terms of the frequency of the most frequent allele are different, the allele frequencies that maximize them are identical. Moreover, we show using coalescent simulations that when taking into account the specific maximal values of the three statistics, their behaviours become similar across a large range of migration rates. We use our results to explain two empirical patterns: the similar values of the three statistics among North American wolves, and the low D values compared to and FST in Atlantic salmon. The results suggest that the three statistics are often predictably similar, so that they can make quite similar contributions to data analysis. When they are not similar, the difference can be understood in relation to features of genetic diversity.
Keywords: allele frequency; gene flow; genetic differentiation; migration; population structure.
© 2019 John Wiley & Sons Ltd.
Figures






References
-
- Beaumont MA, & Nichols RA (1996). Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London. Series B: Biological Sciences, 263, 1619–1626.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

