Altered γ-Secretase Processing of APP Disrupts Lysosome and Autophagosome Function in Monogenic Alzheimer's Disease
- PMID: 30590039
- PMCID: PMC6315085
- DOI: 10.1016/j.celrep.2018.11.095
Altered γ-Secretase Processing of APP Disrupts Lysosome and Autophagosome Function in Monogenic Alzheimer's Disease
Abstract
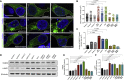
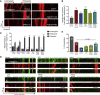
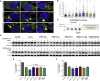
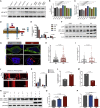
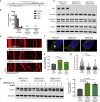
Abnormalities of the endolysosomal and autophagy systems are found in Alzheimer's disease, but it is not clear whether defects in these systems are a cause or consequence of degenerative processes in the disease. In human neuronal models of monogenic Alzheimer's disease, APP and PSEN1 mutations disrupt lysosome function and autophagy, leading to impaired lysosomal proteolysis and defective autophagosome clearance. Processing of APP by γ-secretase is central to the pathogenic changes in the lysosome-autophagy system caused by PSEN1 and APP mutations: reducing production of C-terminal APP by inhibition of BACE1 rescued these phenotypes in both APP and PSEN1 mutant neurons, whereas inhibition of γ-secretase induced lysosomal and autophagic pathology in healthy neurons. Defects in lysosomes and autophagy due to PSEN1 mutations are rescued by CRISPR-knockout of APP. These data demonstrate a key role for proteolysis of the C-terminal of APP by γ-secretase in neuronal dysfunction in monogenic Alzheimer's disease.
Keywords: Alzheimer’s disease; autophagy; axonal transport; endosome; live-cell imaging; lysosome.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bertram L., Tanzi R.E. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat. Rev. Neurosci. 2008;9:768–778. - PubMed
-
- Cataldo A.M., Peterhoff C.M., Troncoso J.C., Gomez-Isla T., Hyman B.T., Nixon R.A. Endocytic pathway abnormalities precede amyloid β deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am. J. Pathol. 2000;157:277–286. - PMC - PubMed
-
- Cataldo A., Rebeck G.W., Ghetri B., Hulette C., Lippa C., Van Broeckhoven C., van Duijn C., Cras P., Bogdanovic N., Bird T. Endocytic disturbances distinguish among subtypes of Alzheimer’s disease and related disorders. Ann. Neurol. 2001;50:661–665. - PubMed
-
- Cataldo A.M., Mathews P.M., Boiteau A.B., Hassinger L.C., Peterhoff C.M., Jiang Y., Mullaney K., Neve R.L., Gruenberg J., Nixon R.A. Down syndrome fibroblast model of Alzheimer-related endosome pathology: accelerated endocytosis promotes late endocytic defects. Am. J. Pathol. 2008;173:370–384. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

