Identification of epistasis loci underlying rice flowering time by controlling population stratification and polygenic effect
- PMID: 30590457
- PMCID: PMC6476725
- DOI: 10.1093/dnares/dsy043
Identification of epistasis loci underlying rice flowering time by controlling population stratification and polygenic effect
Erratum in
-
Identification of epistasis loci underlying rice flowering time by controlling population stratification and polygenic effect.DNA Res. 2019 Apr 1;26(2):193. doi: 10.1093/dnares/dsz007. DNA Res. 2019. PMID: 31222259 Free PMC article. No abstract available.
Abstract
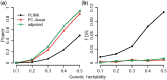
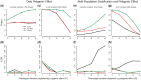
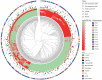
Flowering time is an important agronomic trait, attributed by multiple genes, gene-gene interactions and environmental factors. Population stratification and polygenic effects might confound genetic effects of the causal loci underlying this complex trait. We proposed a two-step approach for detecting epistasis interactions underlying rice flowering time by accounting population structure and polygenic effects. Simulation studies showed that the approach used in this study performs better than classical and PC-linear approaches in terms of powers and false discovery rates in the case of population stratification and polygenic effects. Whole genome epistasis analyses identified 589 putative genetic interactions for flowering time. Eighteen of these interactions are located within 10 kilobases of regions of known protein-protein interactions. Thirty-seven SNPs near to twenty-five genes involve in rice or/and Arabidopsis (orthologue) flowering pathway. Bioinformatics analysis showed that 66.55% pairwise genes of the identified interactions (392 out of the 589 interactions) have similarity in various genomic features. Moreover, significant numbers of detected epistatic genes have high expression in different floral tissues. Our findings highlight the importance of epistasis analysis by controlling population stratification and polygenic effect and provided novel insights into the genetic architecture of rice flowering which could assist breeding programmes.
Keywords: GWAS; epistasis analysis; polygenic effect; population stratification; rice flowering.
© The Author(s) 2018. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures



References
-
- Furbank R.T., von Caemmerer S., Sheehy J., Edwards G.. 2009, C-4 rice: a challenge for plant phenomics, Funct. Plant Biol., 36, 845–56. - PubMed
-
- Roux F., Touzet P., Cuguen J., Le Corre V.. 2006, How to be early flowering: an evolutionary perspective, Trends Plant Sci., 11, 375–81. - PubMed
-
- Yu J., Pressoir G., Briggs W.H., et al.2006, A unified mixed-model method for association mapping that accounts for multiple levels of relatedness, Nat. Genet., 38, 203–8. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

