Genome rearrangements in Escherichia coli during de novo acquisition of resistance to a single antibiotic or two antibiotics successively
- PMID: 30591014
- PMCID: PMC6307192
- DOI: 10.1186/s12864-018-5353-y
Genome rearrangements in Escherichia coli during de novo acquisition of resistance to a single antibiotic or two antibiotics successively
Abstract
Background: The ability of bacteria to acquire resistance to antibiotics relies to a large extent on their capacity for genome modification. Prokaryotic genomes are highly plastic and can utilize horizontal gene transfer, point mutations, and gene deletions or amplifications to realize genome expansion and rearrangements. The contribution of point mutations to de novo acquisition of antibiotic resistance is well-established. In this study, the internal genome rearrangement of Escherichia coli during to de novo acquisition of antibiotic resistance was investigated using whole-genome sequencing.
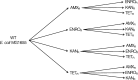
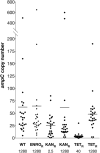
Results: Cells were made resistant to one of the four antibiotics and subsequently to one of the three remaining. This way the initial genetic rearrangements could be documented together with the effects of an altered genetic background on subsequent development of resistance. A DNA fragment including ampC was amplified by a factor sometimes exceeding 100 as a result of exposure to amoxicillin. Excision of prophage e14 was observed in many samples with a double exposure history, but not in cells exposed to a single antibiotic, indicating that the activation of the SOS stress response alone, normally the trigger for excision, was not sufficient to cause excision of prophage e14. Partial deletion of clpS and clpA occurred in strains exposed to enrofloxacin and tetracycline. Other deletions were observed in some strains, but not in replicates with the exact same exposure history. Various insertion sequence transpositions correlated with exposure to specific antibiotics.
Conclusions: Many of the genome rearrangements have not been reported before to occur during resistance development. The observed correlation between genome rearrangements and specific antibiotic pressure, as well as their presence in independent replicates indicates that these events do not occur randomly. Taken together, the observed genome rearrangements illustrate the plasticity of the E. coli genome when exposed to antibiotic stress.
Keywords: Gene amplification; Genome rearrangement; Prophage; de novo resistance.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Effects of a previously selected antibiotic resistance on mutations acquired during development of a second resistance in Escherichia coli.BMC Genomics. 2019 Apr 11;20(1):284. doi: 10.1186/s12864-019-5648-7. BMC Genomics. 2019. PMID: 30975082 Free PMC article.
-
Interaction between mutations and regulation of gene expression during development of de novo antibiotic resistance.Antimicrob Agents Chemother. 2014 Aug;58(8):4371-9. doi: 10.1128/AAC.02892-14. Epub 2014 May 19. Antimicrob Agents Chemother. 2014. PMID: 24841263 Free PMC article.
-
Evaluation of Acquired Antibiotic Resistance in Escherichia coli Exposed to Long-Term Low-Shear Modeled Microgravity and Background Antibiotic Exposure.mBio. 2019 Jan 15;10(1):e02637-18. doi: 10.1128/mBio.02637-18. mBio. 2019. PMID: 30647159 Free PMC article.
-
The mar regulon: multiple resistance to antibiotics and other toxic chemicals.Trends Microbiol. 1999 Oct;7(10):410-3. doi: 10.1016/s0966-842x(99)01589-9. Trends Microbiol. 1999. PMID: 10498949 Review.
-
Multiple Pathways of Genome Plasticity Leading to Development of Antibiotic Resistance.Antibiotics (Basel). 2013 May 30;2(2):288-315. doi: 10.3390/antibiotics2020288. Antibiotics (Basel). 2013. PMID: 27029305 Free PMC article. Review.
Cited by
-
Reactive oxygen species accelerate de novo acquisition of antibiotic resistance in E. coli.iScience. 2023 Oct 31;26(12):108373. doi: 10.1016/j.isci.2023.108373. eCollection 2023 Dec 15. iScience. 2023. PMID: 38025768 Free PMC article.
-
De novo acquisition of antibiotic resistance in six species of bacteria.Microbiol Spectr. 2025 Mar 4;13(3):e0178524. doi: 10.1128/spectrum.01785-24. Epub 2025 Feb 5. Microbiol Spectr. 2025. PMID: 39907470 Free PMC article.
-
Effects of a previously selected antibiotic resistance on mutations acquired during development of a second resistance in Escherichia coli.BMC Genomics. 2019 Apr 11;20(1):284. doi: 10.1186/s12864-019-5648-7. BMC Genomics. 2019. PMID: 30975082 Free PMC article.
-
Role of RelA-synthesized (p)ppGpp and ROS-induced mutagenesis in de novo acquisition of antibiotic resistance in E. coli.iScience. 2024 Mar 26;27(4):109579. doi: 10.1016/j.isci.2024.109579. eCollection 2024 Apr 19. iScience. 2024. PMID: 38617560 Free PMC article.
-
A novel decoy strategy for polymyxin resistance in Acinetobacter baumannii.Elife. 2021 Jun 28;10:e66988. doi: 10.7554/eLife.66988. Elife. 2021. PMID: 34180396 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

