Discerning novel splice junctions derived from RNA-seq alignment: a deep learning approach
- PMID: 30591034
- PMCID: PMC6307148
- DOI: 10.1186/s12864-018-5350-1
Discerning novel splice junctions derived from RNA-seq alignment: a deep learning approach
Abstract
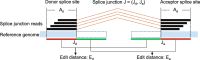
Background: Exon splicing is a regulated cellular process in the transcription of protein-coding genes. Technological advancements and cost reductions in RNA sequencing have made quantitative and qualitative assessments of the transcriptome both possible and widely available. RNA-seq provides unprecedented resolution to identify gene structures and resolve the diversity of splicing variants. However, currently available ab initio aligners are vulnerable to spurious alignments due to random sequence matches and sample-reference genome discordance. As a consequence, a significant set of false positive exon junction predictions would be introduced, which will further confuse downstream analyses of splice variant discovery and abundance estimation.
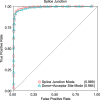
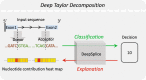
Results: In this work, we present a deep learning based splice junction sequence classifier, named DeepSplice, which employs convolutional neural networks to classify candidate splice junctions. We show (I) DeepSplice outperforms state-of-the-art methods for splice site classification when applied to the popular benchmark dataset HS3D, (II) DeepSplice shows high accuracy for splice junction classification with GENCODE annotation, and (III) the application of DeepSplice to classify putative splice junctions generated by Rail-RNA alignment of 21,504 human RNA-seq data significantly reduces 43 million candidates into around 3 million highly confident novel splice junctions.
Conclusions: A model inferred from the sequences of annotated exon junctions that can then classify splice junctions derived from primary RNA-seq data has been implemented. The performance of the model was evaluated and compared through comprehensive benchmarking and testing, indicating a reliable performance and gross usability for classifying novel splice junctions derived from RNA-seq alignment.
Keywords: Deep learning; Exon splicing; RNA-seq; Splice junction.
Conflict of interest statement
Ethics approval and consent to participate
No permission was required from the ethics committee as the project did not involve testing of human, animal or endangered plant species subjects.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures







Similar articles
-
Discover hidden splicing variations by mapping personal transcriptomes to personal genomes.Nucleic Acids Res. 2015 Dec 15;43(22):10612-22. doi: 10.1093/nar/gkv1099. Epub 2015 Nov 17. Nucleic Acids Res. 2015. PMID: 26578562 Free PMC article.
-
Using RNA-Seq to Discover Genetic Polymorphisms That Produce Hidden Splice Variants.Methods Mol Biol. 2017;1648:129-142. doi: 10.1007/978-1-4939-7204-3_10. Methods Mol Biol. 2017. PMID: 28766294
-
Simulation-based comprehensive benchmarking of RNA-seq aligners.Nat Methods. 2017 Feb;14(2):135-139. doi: 10.1038/nmeth.4106. Epub 2016 Dec 12. Nat Methods. 2017. PMID: 27941783 Free PMC article.
-
Overview of available methods for diverse RNA-Seq data analyses.Sci China Life Sci. 2011 Dec;54(12):1121-8. doi: 10.1007/s11427-011-4255-x. Epub 2012 Jan 7. Sci China Life Sci. 2011. PMID: 22227904 Review.
-
Alternative splicing, RNA-seq and drug discovery.Drug Discov Today. 2019 Jun;24(6):1258-1267. doi: 10.1016/j.drudis.2019.03.030. Epub 2019 Apr 4. Drug Discov Today. 2019. PMID: 30953866 Review.
Cited by
-
A hybrid approach of ensemble learning and grey wolf optimizer for DNA splice junction prediction.PLoS One. 2024 Sep 23;19(9):e0310698. doi: 10.1371/journal.pone.0310698. eCollection 2024. PLoS One. 2024. PMID: 39312561 Free PMC article.
-
Splice Junction Identification using Long Short-Term Memory Neural Networks.Curr Genomics. 2021 Dec 30;22(5):384-390. doi: 10.2174/1389202922666211011143008. Curr Genomics. 2021. PMID: 35283668 Free PMC article.
-
Reference-informed prediction of alternative splicing and splicing-altering mutations from sequences.bioRxiv [Preprint]. 2024 Apr 8:2024.03.22.586363. doi: 10.1101/2024.03.22.586363. bioRxiv. 2024. Update in: Genome Res. 2024 Aug 20;34(7):1052-1065. doi: 10.1101/gr.279044.124. PMID: 38586002 Free PMC article. Updated. Preprint.
-
A benchmark study of ab initio gene prediction methods in diverse eukaryotic organisms.BMC Genomics. 2020 Apr 9;21(1):293. doi: 10.1186/s12864-020-6707-9. BMC Genomics. 2020. PMID: 32272892 Free PMC article.
-
IUP-BERT: Identification of Umami Peptides Based on BERT Features.Foods. 2022 Nov 21;11(22):3742. doi: 10.3390/foods11223742. Foods. 2022. PMID: 36429332 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

