RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer
- PMID: 30591054
- PMCID: PMC6307166
- DOI: 10.1186/s13046-018-1006-x
RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer
Abstract
Background: Circular RNA (circRNA) is a novel class of noncoding RNAs with functions in various pathophysiological activities. However, the expression profiles and functions of circRNAs in colorectal cancer (CRC) remain largely unknown.
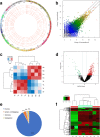
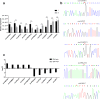
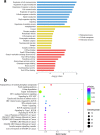
Methods: High-throughput RNA sequencing (RNA-seq) was performed to assess circRNA expression profiles in 4 paired CRC tissues, and significantly dysregulated circRNAs were validated by quantitative real-time polymerase chain reaction (qRT-PCR). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to predict the potential functions of dysregulated circRNAs. Target miRNAs of circRNAs were predicted using miRanda software, and were further analyzed combining DIANA-miRPath v.3 platform (Reverse Search module) with KEGG pathways of COLORECTAL CANCER and MicroRNAs in cancer (Entry: map05210 and map05206). CircRNA-miRNA interaction networks were constructed using Cytoscape software. Expression levels of a significantly down-regulated circRNA, circDDX17 (hsa_circ_0002211), was detected by qRT-PCR in 60 paired CRC tissues. CircDDX17 was knockdown by siRNA, and the biological functions of circDDX17 were examined in CRC cell lines.
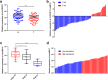
Results: Totally 448 differentially expressed circRNAs were identified, including 394 up-regulated and 54 down-regulated circRNAs. qRT-PCR validation confirmed the reliability of the RNA-Seq data. GO and KEGG analyses revealed that these dysregulated circRNAs were potentially implicated in CRC pathogenesis. Analyses by combining miRanda and miRPath softwares with KEGG pathways suggested that the miRNAs targeted by the top 10 dysregulated circRNAs were associated with the KEGG pathways of COLORECTAL CANCER and MicroRNAs in cancer, indicating that circRNA-miRNA interactions might play important functional roles in the initiation and progression of CRC. The results of qRT-PCR for circDDX17 in 60 paired CRC tissues showed that circDDX17 was significantly down-regulated in CRC tissues and associated with unfavorable clinicopathological parameters. In vitro experiments showed that silencing of circDDX17 promoted CRC cell proliferation, migration, invasion, and inhibited apoptosis.
Conclusions: In conclusion, we have identified numerous circRNAs that are dysregulated in CRC tissues compared with adjacent normal mucosa tissues. Bioinformatic analyses suggested that these dysregulated circRNAs might play important functional roles in CRC tumorigenesis. CircDDX17 functions as a tumor suppressor and could serve as a potential biomarker and a therapeutic target for CRC.
Keywords: Bioinformatic analysis; CircDDX17; Circular RNAs; Colorectal cancer; High-throughput sequencing; Tumor suppressor.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University and conducted in accordance with the ethical standards formulated in the Declaration of Helsinki. Written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

