R/qtl2: Software for Mapping Quantitative Trait Loci with High-Dimensional Data and Multiparent Populations
- PMID: 30591514
- PMCID: PMC6366910
- DOI: 10.1534/genetics.118.301595
R/qtl2: Software for Mapping Quantitative Trait Loci with High-Dimensional Data and Multiparent Populations
Abstract
R/qtl2 is an interactive software environment for mapping quantitative trait loci (QTL) in experimental populations. The R/qtl2 software expands the scope of the widely used R/qtl software package to include multiparent populations derived from more than two founder strains, such as the Collaborative Cross and Diversity Outbred mice, heterogeneous stocks, and MAGIC plant populations. R/qtl2 is designed to handle modern high-density genotyping data and high-dimensional molecular phenotypes, including gene expression and proteomics. R/qtl2 includes the ability to perform genome scans using a linear mixed model to account for population structure, and also includes features to impute SNPs based on founder strain genomes and to carry out association mapping. The R/qtl2 software provides all of the basic features needed for QTL mapping, including graphical displays and summary reports, and it can be extended through the creation of add-on packages. R/qtl2, which is free and open source software written in the R and C++ programming languages, comes with a test framework.
Keywords: Collaborative Cross; Diversity Outbred mice; MAGIC; MPP; Multiparent Advanced Generation Inter-Cross (MAGIC); QTL; heterogeneous stock; multiparent populations; software.
Copyright © 2019 Broman et al.
Figures
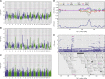
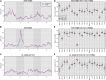
References
-
- Arends D., Li Y., Brockmann G. A., Jansen R. C., Williams R. W., et al. , 2016. Correlation trait loci (CTL) mapping: phenotype network inference subject to genotype. J. Open Source Softw. 1: 87 10.21105/joss.00087 - DOI
-
- Basten C. J., Weir B. S., Zeng Z.-B., 2002. QTL Cartographer, Version 1.16. Department of Statistics, North Carolina State University, Raleigh, NC.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

