Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints
- PMID: 30591526
- PMCID: PMC6405336
- DOI: 10.1182/blood-2018-08-862292
Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints
Abstract
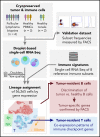
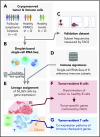
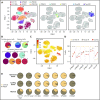
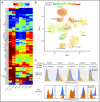
Follicular lymphoma (FL) is a low-grade B-cell malignancy that transforms into a highly aggressive and lethal disease at a rate of 2% per year. Perfect isolation of the malignant B-cell population from a surgical biopsy is a significant challenge, masking important FL biology, such as immune checkpoint coexpression patterns. To resolve the underlying transcriptional networks of follicular B-cell lymphomas, we analyzed the transcriptomes of 34 188 cells derived from 6 primary FL tumors. For each tumor, we identified normal immune subpopulations and malignant B cells, based on gene expression. We used multicolor flow cytometry analysis of the same tumors to confirm our assignments of cellular lineages and validate our predictions of expressed proteins. Comparison of gene expression between matched malignant and normal B cells from the same patient revealed tumor-specific features. Malignant B cells exhibited restricted immunoglobulin (Ig) light chain expression (either Igκ or Igλ), as well the expected upregulation of the BCL2 gene, but also downregulation of the FCER2, CD52, and major histocompatibility complex class II genes. By analyzing thousands of individual cells per patient tumor, we identified the mosaic of malignant B-cell subclones that coexist within a FL and examined the characteristics of tumor-infiltrating T cells. We identified genes coexpressed with immune checkpoint molecules, such as CEBPA and B2M in regulatory T (Treg) cells, providing a better understanding of the gene networks involved in immune regulation. In summary, parallel measurement of single-cell expression in thousands of tumor cells and tumor-infiltrating lymphocytes can be used to obtain a systems-level view of the tumor microenvironment and identify new avenues for therapeutic development.
© 2019 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: G.X.Y.Z. is an employee of 10X Genomics. The remaining authors declare no competing financial interests.
Figures


References
-
- Casulo C, Burack WR, Friedberg JW. Transformed follicular non-Hodgkin lymphoma. Blood. 2015;125(1):40-47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

