Biosynthesis of O-N-acetylgalactosamine glycans in the human cell nucleus
- PMID: 30591584
- PMCID: PMC6398145
- DOI: 10.1074/jbc.RA118.005524
Biosynthesis of O-N-acetylgalactosamine glycans in the human cell nucleus
Erratum in
-
Correction: Biosynthesis of O-N-acetylgalactosamine glycans in the human cell nucleus.J Biol Chem. 2019 May 10;294(19):7967. doi: 10.1074/jbc.AAC119.008943. J Biol Chem. 2019. PMID: 31076524 Free PMC article. No abstract available.
Abstract
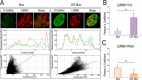
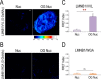
Biological functions of nuclear proteins are regulated by post-translational modifications (PTMs) that modulate gene expression and cellular physiology. However, the role of O-linked glycosylation (O-GalNAc) as a PTM of nuclear proteins in the human cell has not been previously reported. Here, we examined in detail the initiation of O-GalNAc glycan biosynthesis, representing a novel PTM of nuclear proteins in the nucleus of human cells, with an emphasis on HeLa cells. Using soluble nuclear fractions from purified nuclei, enzymatic assays, fluorescence microscopy, affinity chromatography, MS, and FRET analyses, we identified all factors required for biosynthesis of O-GalNAc glycans in nuclei: the donor substrate (UDP-GalNAc), nuclear polypeptide GalNAc -transferase activity, and a GalNAc transferase (polypeptide GalNAc-T3). Moreover, we identified O-GalNAc glycosylated proteins in the nucleus and present solid evidence for O-GalNAc glycan synthesis in this organelle. The demonstration of O-GalNAc glycosylation of nuclear proteins in mammalian cells reported here has important implications for cell and chemical biology.
Keywords: O-GalNAc glycans; biosynthesis; enzyme catalysis; epigenetics; glycobiology; glycoconjugate; glycoprotein biosynthesis; glycosylation; nucleus; polypeptide N-acetylgalactosaminyltransferase 3; post-translational modification; proteins; substrate specificity.
© 2019 Cejas et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

