Quantifying site-specific chromatin mechanics and DNA damage response
- PMID: 30591710
- PMCID: PMC6308236
- DOI: 10.1038/s41598-018-36343-x
Quantifying site-specific chromatin mechanics and DNA damage response
Abstract
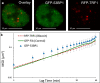
DNA double-strand breaks pose a direct threat to genomic stability. Studies of DNA damage and chromatin dynamics have yielded opposing results that support either increased or decreased chromatin motion after damage. In this study, we independently measure the dynamics of transcriptionally active or repressed chromatin regions using particle tracking microrheology. We find that the baseline motion of transcriptionally repressed regions of chromatin are significantly less mobile than transcriptionally active chromatin, which is statistically similar to the bulk motion of chromatin within the nucleus. Site specific DNA damage using KillerRed tags induced in loci within repressed chromatin causes an increased motion, while loci within transcriptionally active regions remains unchanged at similar time scales. We also observe a time-dependent response associated with a further increase in chromatin decondensation. Global induction of damage with bleocin displays similar trends of chromatin decondensation and increased mobility only at 53BP1-labeled damage sites but not at non-damaged sites, indicating that chromatin dynamics are tightly regulated locally after damage. These results shed light on the evolution of the local and global DNA damage response associated with chromatin remodeling and dynamics, with direct implications for their role in repair.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Transcriptional regulation and chromatin dynamics at DNA double-strand breaks.Exp Mol Med. 2022 Oct;54(10):1705-1712. doi: 10.1038/s12276-022-00862-5. Epub 2022 Oct 13. Exp Mol Med. 2022. PMID: 36229590 Free PMC article. Review.
-
Chromosome Dynamics in Response to DNA Damage.Annu Rev Genet. 2018 Nov 23;52:295-319. doi: 10.1146/annurev-genet-120417-031334. Epub 2018 Sep 12. Annu Rev Genet. 2018. PMID: 30208290 Review.
-
Chromatin mobility is increased at sites of DNA double-strand breaks.J Cell Sci. 2012 May 1;125(Pt 9):2127-33. doi: 10.1242/jcs.089847. Epub 2012 Feb 10. J Cell Sci. 2012. PMID: 22328517
-
Chromatin stiffening underlies enhanced locus mobility after DNA damage in budding yeast.EMBO J. 2017 Sep 1;36(17):2595-2608. doi: 10.15252/embj.201695842. Epub 2017 Jul 10. EMBO J. 2017. PMID: 28694242 Free PMC article.
-
Clustered DNA damage concentrated in particle trajectories causes persistent large-scale rearrangements in chromatin architecture.Radiother Oncol. 2018 Dec;129(3):600-610. doi: 10.1016/j.radonc.2018.07.003. Epub 2018 Jul 23. Radiother Oncol. 2018. PMID: 30049456
Cited by
-
Site-specific targeting of a light activated dCas9-KillerRed fusion protein generates transient, localized regions of oxidative DNA damage.PLoS One. 2020 Dec 17;15(12):e0237759. doi: 10.1371/journal.pone.0237759. eCollection 2020. PLoS One. 2020. PMID: 33332350 Free PMC article.
-
Diffusion controls local versus dispersed inheritance of histones during replication and shapes epigenomic architecture.PLoS Comput Biol. 2023 Dec 18;19(12):e1011725. doi: 10.1371/journal.pcbi.1011725. eCollection 2023 Dec. PLoS Comput Biol. 2023. PMID: 38109423 Free PMC article.
-
Mechanical Forces in Nuclear Organization.Cold Spring Harb Perspect Biol. 2022 Jan 4;14(1):a039685. doi: 10.1101/cshperspect.a039685. Cold Spring Harb Perspect Biol. 2022. PMID: 34187806 Free PMC article. Review.
-
Apoptotic Potential of Glucomoringin Isothiocyanate (GMG-ITC) Isolated from Moringa oleifera Lam Seeds on Human Prostate Cancer Cells (PC-3).Molecules. 2023 Apr 4;28(7):3214. doi: 10.3390/molecules28073214. Molecules. 2023. PMID: 37049977 Free PMC article.
-
The mechanobiology of nuclear phase separation.APL Bioeng. 2022 Apr 28;6(2):021503. doi: 10.1063/5.0083286. eCollection 2022 Jun. APL Bioeng. 2022. PMID: 35540725 Free PMC article. Review.
References
-
- Spagnol ST, Dahl KN. Active cytoskeletal force and chromatin condensation independently modulate intranuclear network fluctuations. Int. Bio. 2014;6:523–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM118833/GM/NIGMS NIH HHS/United States
- EB003392/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- GM118833/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- CMMI-1300476/National Science Foundation (NSF)/International
LinkOut - more resources
Full Text Sources

