N-Acylated amino acid methyl esters from marine Roseobacter group bacteria
- PMID: 30591820
- PMCID: PMC6296433
- DOI: 10.3762/bjoc.14.276
N-Acylated amino acid methyl esters from marine Roseobacter group bacteria
Abstract
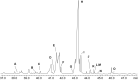
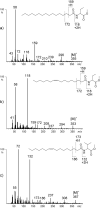
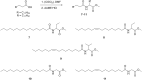
Bacteria of the Roseobacter group (Rhodobacteraceae) are important members of many marine ecosystems. Similar to other Gram-negative bacteria many roseobacters produce N-acylhomoserine lactones (AHLs) for communication by quorum sensing systems. AHLs regulate different traits like cell differentiation or antibiotic production. Related N-acylalanine methyl esters (NAMEs) have been reported as well, but so far only from Roseovarius tolerans EL-164. While screening various roseobacters isolated from macroalgae we encountered four strains, Roseovarius sp. D12_1.68, Loktanella sp. F13, F14 and D3 that produced new derivatives and analogs of NAMEs, namely N-acyl-2-aminobutyric acid methyl esters (NABME), N-acylglycine methyl esters (NAGME), N-acylvaline methyl esters (NAVME), as well as for the first time a methyl-branched NAME, N-(13-methyltetradecanoyl)alanine methyl ester. These compounds were detected by GC-MS analysis, and structural proposals were derived from the mass spectra and by derivatization. Verification of compound structures was performed by synthesis. NABMEs, NAVMEs and NAGMEs are produced in low amounts only, making mass spectrometry the method of choice for their detection. The analysis of both EI and ESI mass spectra revealed fragmentation patterns helpful for the detection of similar compounds derived from other amino acids. Some of these compounds showed antimicrobial activity. The structural similarity of N-acylated amino acid methyl esters and similar lipophilicity to AHLs might indicate a yet unknown function as signalling compounds in the ecology of these bacteria, although their singular occurrence is in strong contrast to the common occurrence of AHLs. Obviously the structural motif is not restricted to Roseovarius spp. and occurs also in other genera.
Keywords: 2-aminobutyric acid; amino acid derivatives; homoserine lactones; natural products; quorum sensing.
Figures
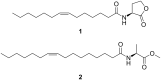

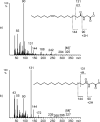
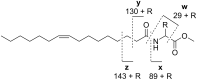
 -alanine methyl ester.
-alanine methyl ester.


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
