A whole genome approach for discovering the genetic basis of blood group antigens: independent confirmation for P1 and Xga
- PMID: 30592300
- PMCID: PMC6402986
- DOI: 10.1111/trf.15089
A whole genome approach for discovering the genetic basis of blood group antigens: independent confirmation for P1 and Xga
Abstract
Background: Although P1 and Xga are known to be associated with the A4GALT and XG genes, respectively, the genetic basis of antigen expression has been elusive. Recent reports link both P1 and Xga expression with nucleotide changes in the promotor regions and with antigen-negative phenotypes due to disruption of transcription factor binding.
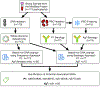
Study design and methods: Whole genome sequencing was performed on 113 individuals as part of the MedSeq Project with serologic RBC antigen typing for P1 (n = 77) and Xga (n = 15). Genomic data were analyzed by two approaches, nucleotide frequency correlation and serologic correlation, to find A4GALT and XG changes associated with P1 and Xga expression.
Results: For P1, the frequency approach identified 29 possible associated nucleotide changes, and the serologic approach revealed four among them correlating with the P1+/P1- phenotype: chr22:43,115,523_43,115,520AAAG/delAAAG (rs66781836); chr 22:43,114,551C/T (rs8138197); chr22:43,114,020 T/G (rs2143918); and chr22:43,113,793G/T (rs5751348). For Xga , the frequency approach identified 82 possible associated nucleotide changes, and among these the serologic approach revealed one correlating with the Xg(a+)/Xg(a-) phenotype: chrX:2,666,384G/C (rs311103).
Conclusion: A bioinformatics analysis pipeline was created to identify genetic changes responsible for RBC antigen expression. This study, in progress before the recently published reports, independently confirms the basis for P1 and Xga . Although this enabled molecular typing of these antigens, the Y chromosome PAR1 region interfered with Xga typing in males. This approach could be used to identify and confirm the genetic basis of antigens, potentially replacing the historical approach using family pedigrees as genomic sequencing becomes commonplace.
© 2018 AABB.
Conflict of interest statement
Declaration of Interests
All authors declare that they have no conflicts of interest relevant to the manuscript.
Figures


References
-
- Westman JS, Stenfelt L, Vidovic K, Moller M, Hellberg A, Kjellstrom S, Olsson ML. Allele-selective RUNX1 binding regulates P1 blood group status by transcriptional control of A4GALT. Blood 2018;131: 1611–6. - PubMed
-
- Moller M, Lee YQ, Vidovic K, Kjellstrom S, Bjorkman L, Storry JR, Olsson ML. Disruption of a GATA1-binding motif upstream of XG/PBDX abolishes Xg(a) expression and resolves the Xg blood group system. Blood 2018;132: 334–8. - PubMed
-
- Landsteiner K, Levine P. Further Observations on Individual Differences of Human Blood. Experimental Biology and Medicine 1927;24: 941–2.
-
- Steffensen R, Carlier K, Wiels J, Levery SB, Stroud M, Cedergren B, Nilsson Sojka B, Bennett EP, Jersild C, Clausen H. Cloning and expression of the histo-blood group Pk UDP-galactose: Ga1beta-4G1cbeta1-cer alpha1, 4-galactosyltransferase. Molecular genetic basis of the p phenotype. J Biol Chem 2000;275: 16723–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

