Biomarkers from circulating neutrophil transcriptomes have potential to detect unruptured intracranial aneurysms
- PMID: 30593281
- PMCID: PMC6310942
- DOI: 10.1186/s12967-018-1749-3
Biomarkers from circulating neutrophil transcriptomes have potential to detect unruptured intracranial aneurysms
Abstract
Background: Intracranial aneurysms (IAs) are dangerous because of their potential to rupture and cause deadly subarachnoid hemorrhages. Previously, we found significant RNA expression differences in circulating neutrophils between patients with unruptured IAs and aneurysm-free controls. Searching for circulating biomarkers for unruptured IAs, we tested the feasibility of developing classification algorithms that use neutrophil RNA expression levels from blood samples to predict the presence of an IA.
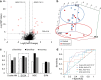
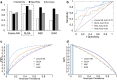
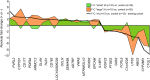
Methods: Neutrophil RNA extracted from blood samples from 40 patients (20 with angiography-confirmed unruptured IA, 20 angiography-confirmed IA-free controls) was subjected to next-generation RNA sequencing to obtain neutrophil transcriptomes. In a randomly-selected training cohort of 30 of the 40 samples (15 with IA, 15 controls), we performed differential expression analysis. Significantly differentially expressed transcripts (false discovery rate < 0.05, fold change ≥ 1.5) were used to construct prediction models for IA using four well-known supervised machine-learning approaches (diagonal linear discriminant analysis, cosine nearest neighbors, nearest shrunken centroids, and support vector machines). These models were tested in a testing cohort of the remaining 10 neutrophil samples from the 40 patients (5 with IA, 5 controls), and model performance was assessed by receiver-operating-characteristic (ROC) curves. Real-time quantitative polymerase chain reaction (PCR) was used to corroborate expression differences of a subset of model transcripts in neutrophil samples from a new, separate validation cohort of 10 patients (5 with IA, 5 controls).
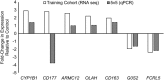
Results: The training cohort yielded 26 highly significantly differentially expressed neutrophil transcripts. Models using these transcripts identified IA patients in the testing cohort with accuracy ranging from 0.60 to 0.90. The best performing model was the diagonal linear discriminant analysis classifier (area under the ROC curve = 0.80 and accuracy = 0.90). Six of seven differentially expressed genes we tested were confirmed by quantitative PCR using isolated neutrophils from the separate validation cohort.
Conclusions: Our findings demonstrate the potential of machine-learning methods to classify IA cases and create predictive models for unruptured IAs using circulating neutrophil transcriptome data. Future studies are needed to replicate these findings in larger cohorts.
Keywords: Inflammation; Intracranial aneurysm; Machine learning; Neutrophils; Transcriptomics.
Figures

Similar articles
-
Classification models using circulating neutrophil transcripts can detect unruptured intracranial aneurysm.J Transl Med. 2020 Oct 15;18(1):392. doi: 10.1186/s12967-020-02550-2. J Transl Med. 2020. PMID: 33059716 Free PMC article.
-
Circulating neutrophil transcriptome may reveal intracranial aneurysm signature.PLoS One. 2018 Jan 17;13(1):e0191407. doi: 10.1371/journal.pone.0191407. eCollection 2018. PLoS One. 2018. PMID: 29342213 Free PMC article.
-
Whole blood transcriptome biomarkers of unruptured intracranial aneurysm.PLoS One. 2020 Nov 6;15(11):e0241838. doi: 10.1371/journal.pone.0241838. eCollection 2020. PLoS One. 2020. PMID: 33156839 Free PMC article.
-
Non-Coding RNAs as Circulating Biomarkers for the Diagnosis of Intracranial Aneurysm: A Systematic Review and Meta-Analysis.J Stroke Cerebrovasc Dis. 2021 Jun;30(6):105762. doi: 10.1016/j.jstrokecerebrovasdis.2021.105762. Epub 2021 Apr 1. J Stroke Cerebrovasc Dis. 2021. PMID: 33813080
-
Transcriptomic Studies on Intracranial Aneurysms.Genes (Basel). 2023 Feb 28;14(3):613. doi: 10.3390/genes14030613. Genes (Basel). 2023. PMID: 36980884 Free PMC article. Review.
Cited by
-
A mendelian randomization study investigates the causal relationship between immune cell phenotypes and cerebral aneurysm.Front Genet. 2024 Jan 19;15:1333855. doi: 10.3389/fgene.2024.1333855. eCollection 2024. Front Genet. 2024. PMID: 38313677 Free PMC article.
-
Characterization of Long Non-coding RNA Signatures of Intracranial Aneurysm in Circulating Whole Blood.Mol Diagn Ther. 2020 Dec;24(6):723-736. doi: 10.1007/s40291-020-00494-3. Mol Diagn Ther. 2020. PMID: 32939739
-
Artificial Intelligence Technologies in Neurosurgery: a Systematic Literature Review Using Topic Modeling. Part II: Research Objectives and Perspectives.Sovrem Tekhnologii Med. 2021;12(6):111-118. doi: 10.17691/stm2020.12.6.12. Epub 2020 Dec 28. Sovrem Tekhnologii Med. 2021. PMID: 34796024 Free PMC article.
-
Aspirin treatment for unruptured intracranial aneurysms: Focusing on its anti-inflammatory role.Heliyon. 2024 Apr 4;10(7):e29119. doi: 10.1016/j.heliyon.2024.e29119. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38617958 Free PMC article. Review.
-
Identification of the key immune-related genes in aneurysmal subarachnoid hemorrhage.Front Mol Neurosci. 2022 Sep 12;15:931753. doi: 10.3389/fnmol.2022.931753. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36172261 Free PMC article.
References
-
- Vega C, Kwoon JV, Lavine SD. Intracranial aneurysms: current evidence and clinical practice. Am Fam Physician. 2002;66:601–608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

