Selective profiling of ribosomes associated with yeast Upf proteins
- PMID: 30593864
- PMCID: PMC6387845
- DOI: 10.1016/j.ymeth.2018.12.008
Selective profiling of ribosomes associated with yeast Upf proteins
Abstract
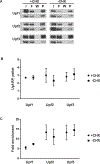
Ribosomes associated with nonsense-mediated decay factors Upf1, Upf2, or Upf3 were purified by immunoprecipitation, and enrichment and stoichiometry of Upf factors and ribosomal proteins were analyzed by western blot and mass spectrometry. Using a small RNA library preparation protocol that eliminates in-gel RNA and cDNA size selection and incorporates four random nucleotides on each side of the ribosome-protected RNA fragment allowed recovery, detection, and analysis of all size classes of protected fragments from a sample simultaneously.
Keywords: NMD; Selective ribosome profiling; Upf proteins.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures


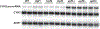

Similar articles
-
Ribosome-bound Upf1 forms distinct 80S complexes and conducts mRNA surveillance.RNA. 2022 Dec;28(12):1621-1642. doi: 10.1261/rna.079416.122. Epub 2022 Oct 3. RNA. 2022. PMID: 36192133 Free PMC article.
-
Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes.EMBO J. 2018 Nov 2;37(21):e99278. doi: 10.15252/embj.201899278. Epub 2018 Oct 1. EMBO J. 2018. PMID: 30275269 Free PMC article.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit.RNA. 2013 Aug;19(8):1105-15. doi: 10.1261/rna.039396.113. Epub 2013 Jun 25. RNA. 2013. PMID: 23801788 Free PMC article.
-
Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives.Biochim Biophys Acta. 2013 Aug;1829(8):775-90. doi: 10.1016/j.bbagrm.2013.01.007. Epub 2013 Jan 26. Biochim Biophys Acta. 2013. PMID: 23357782 Review.
Cited by
-
Features and factors that dictate if terminating ribosomes cause or counteract nonsense-mediated mRNA decay.J Biol Chem. 2022 Nov;298(11):102592. doi: 10.1016/j.jbc.2022.102592. Epub 2022 Oct 13. J Biol Chem. 2022. PMID: 36244451 Free PMC article. Review.
-
Transcriptome-wide investigation of stop codon readthrough in Saccharomyces cerevisiae.PLoS Genet. 2021 Apr 20;17(4):e1009538. doi: 10.1371/journal.pgen.1009538. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33878104 Free PMC article.
-
Identification of the hyaluronic acid pathway as a therapeutic target for facioscapulohumeral muscular dystrophy.Sci Adv. 2019 Dec 11;5(12):eaaw7099. doi: 10.1126/sciadv.aaw7099. eCollection 2019 Dec. Sci Adv. 2019. PMID: 31844661 Free PMC article.
-
Ribosome-bound Upf1 forms distinct 80S complexes and conducts mRNA surveillance.RNA. 2022 Dec;28(12):1621-1642. doi: 10.1261/rna.079416.122. Epub 2022 Oct 3. RNA. 2022. PMID: 36192133 Free PMC article.
References
-
- Nicholson P, Joncourt R, Muhlemann O, Analysis of nonsense-mediated mRNA decay in mammalian cells, Current protocols in cell biology editorial board, Juan S. Bonifacino [et al.] Chapter 27 (2012) Unit27 4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

