Mitochondrial Sco proteins are involved in oxidative stress defense
- PMID: 30593977
- PMCID: PMC6307045
- DOI: 10.1016/j.redox.2018.101079
Mitochondrial Sco proteins are involved in oxidative stress defense
Abstract
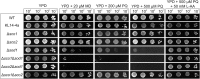
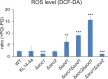
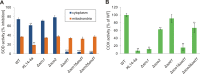
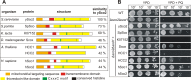
Members of the evolutionary conserved Sco protein family have been intensively studied regarding their role in the assembly of the mitochondrial cytochrome c oxidase. However, experimental and structural data, specifically the presence of a thioredoxin-like fold, suggest that Sco proteins may also play a role in redox homeostasis. In our study, we addressed this putative function of Sco proteins using Saccharomyces cerevisiae as a model system. Like many eukaryotes, this yeast possesses two SCO homologs (SCO1 and SCO2). Mutants bearing a deletion of either of the two genes are not affected in their growth under oxidative stress. However, the concomitant deletion of the SOD1 gene encoding the superoxide dismutase 1 resulted in a distinct phenotype: double deletion strains lacking SCO1 or SCO2 and SOD1 are highly sensitive to oxidative stress and show dramatically increased ROS levels. The respiratory competent double deletion strain Δsco2Δsod1 paved the way to investigate the putative antioxidant function of SCO homologs apart from their role in respiration by complementation analysis. Sco homologs from Drosophila, Arabidopsis, human and two other yeast species were integrated into the genome of the double deletion mutant and the transformants were analyzed for their growth under oxidative stress. Interestingly, all homologs except for Kluyveromyces lactis K07152 and Arabidopsis thaliana HCC1 were able to complement the phenotype, indicating their role in oxidative stress defense. We further applied this complementation-based system to investigate whether pathogenic point mutations affect the putative antioxidant role of hSco2. Surprisingly, all of the mutant alleles failed to restore the ROS-sensitivity of the Δsco2Δsod1 strain. In conclusion, our data not only provide clear evidence for the function of Sco proteins in oxidative stress defense but also offer a valuable tool to investigate this role for other homologous proteins.
Keywords: Mitochondria; Oxidative stress response; ROS; Saccharomyces cerevisiae; Sco proteins; Sod1.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
The mitochondrial copper chaperone COX11 has an additional role in cellular redox homeostasis.PLoS One. 2021 Dec 17;16(12):e0261465. doi: 10.1371/journal.pone.0261465. eCollection 2021. PLoS One. 2021. PMID: 34919594 Free PMC article.
-
HCC1, the Arabidopsis homologue of the yeast mitochondrial copper chaperone SCO1, is essential for embryonic development.J Exp Bot. 2011 Jan;62(1):319-30. doi: 10.1093/jxb/erq269. Epub 2010 Nov 1. J Exp Bot. 2011. PMID: 21041373
-
The role of two putative nitroreductases, Frm2p and Hbn1p, in the oxidative stress response in Saccharomyces cerevisiae.Yeast. 2010 Feb;27(2):89-102. doi: 10.1002/yea.1734. Yeast. 2010. PMID: 19904831
-
Redox regulation of SCO protein function: controlling copper at a mitochondrial crossroad.Antioxid Redox Signal. 2010 Nov 1;13(9):1403-16. doi: 10.1089/ars.2010.3116. Antioxid Redox Signal. 2010. PMID: 20136502 Review.
-
Getting out what you put in: Copper in mitochondria and its impacts on human disease.Biochim Biophys Acta Mol Cell Res. 2021 Jan;1868(1):118867. doi: 10.1016/j.bbamcr.2020.118867. Epub 2020 Oct 2. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 32979421 Free PMC article. Review.
Cited by
-
Biochemistry of Copper Site Assembly in Heme-Copper Oxidases: A Theme with Variations.Int J Mol Sci. 2019 Aug 5;20(15):3830. doi: 10.3390/ijms20153830. Int J Mol Sci. 2019. PMID: 31387303 Free PMC article. Review.
-
Loss of Functional SCO2 Attenuates Oxidative Stress in Diabetic Kidney Disease.Diabetes. 2021 Oct 26;71(1):142-56. doi: 10.2337/db21-0316. Online ahead of print. Diabetes. 2021. PMID: 34702781 Free PMC article.
-
Azoramide protects iPSC-derived dopaminergic neurons with PLA2G6 D331Y mutation through restoring ER function and CREB signaling.Cell Death Dis. 2020 Feb 18;11(2):130. doi: 10.1038/s41419-020-2312-8. Cell Death Dis. 2020. PMID: 32071291 Free PMC article.
-
Recurrent sequence evolution after independent gene duplication.BMC Evol Biol. 2020 Aug 8;20(1):98. doi: 10.1186/s12862-020-01660-1. BMC Evol Biol. 2020. PMID: 32770961 Free PMC article.
-
Development of a novel PTD-mediated IVT-mRNA delivery platform for potential protein replacement therapy of metabolic/genetic disorders.Mol Ther Nucleic Acids. 2021 Sep 20;26:694-710. doi: 10.1016/j.omtn.2021.09.008. eCollection 2021 Dec 3. Mol Ther Nucleic Acids. 2021. PMID: 34703653 Free PMC article.
References
-
- Banci L., Bertini I., Cavallaro G., Rosato A. The functions of Sco proteins from genome-based analysis. J. Proteome Res. 2007;6(4):1568. - PubMed
-
- Banci L., Bertini I., Cavallaro G., Ciofi-Baffoni S. Seeking the determinants of the elusive functions of Sco proteins. FEBS J. 2011;278(13):2244. - PubMed
-
- Schulze M., Rödel G. SCO1, a yeast nuclear gene essential for accumulation of mitochondrial cytochrome c oxidase subunit II. Mol. Gen. Genet. 1988;211(3):492. - PubMed
-
- Krummeck G., Rödel G. Yeast SCO1 protein is required for a post-translational step in the accumulation of mitochondrial cytochrome c oxidase subunits I and II. Curr. Genet. 1990;18(1):13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

