Development and validation of an immune gene-set based Prognostic signature in ovarian cancer
- PMID: 30594555
- PMCID: PMC6412087
- DOI: 10.1016/j.ebiom.2018.12.054
Development and validation of an immune gene-set based Prognostic signature in ovarian cancer
Abstract
Background: Ovarian cancer (OV) is the most lethal gynecological cancer in women. We aim to develop a generalized, individualized immune prognostic signature that can stratify and predict overall survival for ovarian cancer.
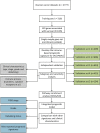
Methods: The gene expression profiles of ovarian cancer tumor tissue samples were collected from 17 public cohorts, including 2777 cases totally. Single sample gene set enrichment (ssGSEA) analysis was used for the immune genes from ImmPort database to develop an immune-based prognostic score for OV (IPSOV). The signature was trained and validated in six independent datasets (n = 519, 409, 606, 634, 415, 194).
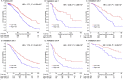
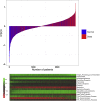
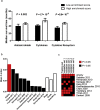
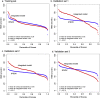
Findings: The IPSOV significantly stratified patients into low- and high-immune risk groups in the training set and in the 5 validation sets (HR range: 1.71 [95%CI: 1.32-2.19; P = 4.04 × 10-5] to 2.86 [95%CI: 1.72-4.74; P = 4.89 × 10-5]). Further, we compared IPSOV with nine reported ovarian cancer prognostic signatures as well as the clinical characteristics including stage, grade and debulking status. The IPSOV achieved the highest mean C-index (0.625) compared with the other signatures (0.516 to 0.602) and clinical characteristics (0.555 to 0.583). Further, we integrated IPSOV with stage, grade and debulking, which showed improved prognostic accuracy than clinical characteristics only.
Interpretation: The proposed clinical-immune signature is a promising biomarker for estimating overall survival in ovarian cancer. Prospective studies are needed to further validate its analytical accuracy and test the clinical utility. FUND: This work was supported by National Key Research and Development Program of China, National Natural Science Foundation of China and Natural Science Foundation of the Jiangsu Higher Education Institutions of China.
Keywords: Gene expression; Immune; Ovarian cancer; Prognostic signature.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non-Small Cell Lung Cancer.JAMA Oncol. 2017 Nov 1;3(11):1529-1537. doi: 10.1001/jamaoncol.2017.1609. JAMA Oncol. 2017. PMID: 28687838 Free PMC article.
-
Development and verification of an immune-related gene pairs prognostic signature in ovarian cancer.J Cell Mol Med. 2021 Mar;25(6):2918-2930. doi: 10.1111/jcmm.16327. Epub 2021 Feb 4. J Cell Mol Med. 2021. PMID: 33543590 Free PMC article.
-
A prognostic model based on immune-related long noncoding RNAs for patients with epithelial ovarian cancer.J Ovarian Res. 2022 Jan 15;15(1):8. doi: 10.1186/s13048-021-00930-w. J Ovarian Res. 2022. PMID: 35031063 Free PMC article.
-
Increased expression of TET3 predicts unfavorable prognosis in patients with ovarian cancer-a bioinformatics integrative analysis.J Ovarian Res. 2019 Oct 27;12(1):101. doi: 10.1186/s13048-019-0575-4. J Ovarian Res. 2019. PMID: 31656201 Free PMC article. Review.
-
miRNA-200a/c as potential biomarker in epithelial ovarian cancer (EOC): evidence based on miRNA meta-signature and clinical investigations.Oncotarget. 2016 Dec 6;7(49):81621-81633. doi: 10.18632/oncotarget.13154. Oncotarget. 2016. PMID: 27835595 Free PMC article. Review.
Cited by
-
Tumor Immunometabolism Characterization in Ovarian Cancer With Prognostic and Therapeutic Implications.Front Oncol. 2021 Mar 16;11:622752. doi: 10.3389/fonc.2021.622752. eCollection 2021. Front Oncol. 2021. PMID: 33796460 Free PMC article.
-
Comprehensive profiling of immune-related genes in soft tissue sarcoma patients.J Transl Med. 2020 Sep 1;18(1):337. doi: 10.1186/s12967-020-02512-8. J Transl Med. 2020. PMID: 32873319 Free PMC article.
-
Interplay of Sphingolipid Metabolism in Predicting Prognosis of GBM Patients: Towards Precision Immunotherapy.J Cancer. 2024 Jan 1;15(1):275-292. doi: 10.7150/jca.89338. eCollection 2024. J Cancer. 2024. PMID: 38164288 Free PMC article.
-
Identification of m6A regulator-mediated RNA methylation modification patterns and key immune-related genes involved in atrial fibrillation.Aging (Albany NY). 2023 Feb 20;15(5):1371-1393. doi: 10.18632/aging.204537. Epub 2023 Feb 20. Aging (Albany NY). 2023. PMID: 36863715 Free PMC article.
-
Pancreatic adenocarcinoma associated immune-gene signature as a novo risk factor for clinical prognosis prediction in hepatocellular carcinoma.Sci Rep. 2022 Jul 13;12(1):11944. doi: 10.1038/s41598-022-16155-w. Sci Rep. 2022. PMID: 35831362 Free PMC article.
References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47(4):320. - PubMed
-
- Ng S.W., Mitchell A., Kennedy J.A., Chen W.C., McLeod J., Ibrahimova N. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

