Electron microscopy combined with spatial analysis: quantitative mapping of the nano-assemblies of plasma membrane-associating proteins and lipids
- PMID: 30596140
- PMCID: PMC6276063
- DOI: 10.1007/s41048-018-0060-4
Electron microscopy combined with spatial analysis: quantitative mapping of the nano-assemblies of plasma membrane-associating proteins and lipids
Abstract
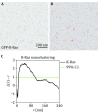
The plasma membrane (PM) is a complex environment consisting of > 700 species of lipids and many different types of membrane-associating proteins. These lipids and membrane proteins are distributed heterogeneously into nanometer-sized domains, called nanoclusters. The lateral spatial segregation in the PM gives rise to different curvature and lipid composition, which determines the efficiency of effector binding and signal transmission. Here, we describe an electron microscopy (EM)-spatial mapping technique to quantify the extent of nanoclusters formation in the PM. The nano-assemblies in the PM are quantified via expressing the GFP-tagged proteins or lipid-binding domains in the cells, which are then immunolabeled with the gold nanoparticles pre-coupled to the anti-GFP antibody. The gold nanoparticles are visualized via the transmission EM at high magnification. The statistical analysis of the Ripley's K-function calculates the spatial distribution of the gold nanoparticles. Important spatial parameters, such as the extent of nanoclustering, the clustered fraction, the number of proteins per cluster, the optimal size of a nanocluster, and the number of proteins localized to the PM, can be calculated. Further detailed aggregation pattern, such as the populations of monomers, dimers, trimers, and higher ordered oligomers, can also be extracted from the spatial analysis. The EM-bivariate analysis quantifies the extent of co-localization between two different components in the PM and provides key information on the protein-protein and the protein-lipid interactions over a long-distance scale from 8 to 240 nm.
Keywords: Electron microscopy; Lipid-anchored proteins; Nanoclustering; Ripley’s K-function; Signal transduction; Spatial distribution.
Conflict of interest statement
Yong Zhou and John F. Hancock declare that they have no conflict of interest.This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures

References
-
- Diggle PJ, Mateu J, Clough HE. A comparison between parametric and non-parametric approaches to the analysis of replicated spatial point patterns. Adv Appl Probab. 2000;32:331–343. doi: 10.1239/aap/1013540166. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
