Enabling precision medicine via standard communication of HTS provenance, analysis, and results
- PMID: 30596645
- PMCID: PMC6338479
- DOI: 10.1371/journal.pbio.3000099
Enabling precision medicine via standard communication of HTS provenance, analysis, and results
Abstract
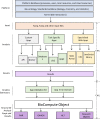
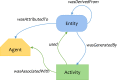
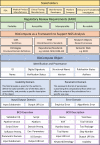
A personalized approach based on a patient's or pathogen's unique genomic sequence is the foundation of precision medicine. Genomic findings must be robust and reproducible, and experimental data capture should adhere to findable, accessible, interoperable, and reusable (FAIR) guiding principles. Moreover, effective precision medicine requires standardized reporting that extends beyond wet-lab procedures to computational methods. The BioCompute framework (https://w3id.org/biocompute/1.3.0) enables standardized reporting of genomic sequence data provenance, including provenance domain, usability domain, execution domain, verification kit, and error domain. This framework facilitates communication and promotes interoperability. Bioinformatics computation instances that employ the BioCompute framework are easily relayed, repeated if needed, and compared by scientists, regulators, test developers, and clinicians. Easing the burden of performing the aforementioned tasks greatly extends the range of practical application. Large clinical trials, precision medicine, and regulatory submissions require a set of agreed upon standards that ensures efficient communication and documentation of genomic analyses. The BioCompute paradigm and the resulting BioCompute Objects (BCOs) offer that standard and are freely accessible as a GitHub organization (https://github.com/biocompute-objects) following the "Open-Stand.org principles for collaborative open standards development." With high-throughput sequencing (HTS) studies communicated using a BCO, regulatory agencies (e.g., Food and Drug Administration [FDA]), diagnostic test developers, researchers, and clinicians can expand collaboration to drive innovation in precision medicine, potentially decreasing the time and cost associated with next-generation sequencing workflow exchange, reporting, and regulatory reviews.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, et al. (2017) LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33: 272–279. 10.1093/bioinformatics/btw613 - DOI - PMC - PubMed
-
- Sawyer E (2017) High Throughput Sequencing and Cost Trends. Nature Education.
-
- Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; Institute of Medicine; Micheel CM, Nass SJ, Omenn GS, editors. (2012). Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

