MMP13 inhibition rescues cognitive decline in Alzheimer transgenic mice via BACE1 regulation
- PMID: 30596903
- PMCID: PMC6657286
- DOI: 10.1093/brain/awy305
MMP13 inhibition rescues cognitive decline in Alzheimer transgenic mice via BACE1 regulation
Abstract
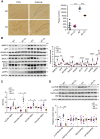
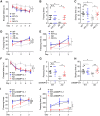
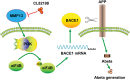
MMP13 (matrix metallopeptidase 13) plays a key role in bone metabolism and cancer development, but has no known functions in Alzheimer's disease. In this study, we used high-throughput small molecule screening in SH-SY5Y cells that stably expressed a luciferase reporter gene driven by the BACE1 (β-site amyloid precursor protein cleaving enzyme 1) promoter, which included a portion of the 5' untranslated region (5'UTR). We identified that CL82198, a selective inhibitor of MMP13, decreased BACE1 protein levels in cultured neuronal cells. This effect was dependent on PI3K (phosphatidylinositide 3-kinase) signalling, and was unrelated to BACE1 gene transcription and protein degradation. Further, we found that eukaryotic translation initiation factor 4B (eIF4B) played a key role, as the mutation of eIF4B at serine 422 (S422R) or deletion of the BACE1 5'UTR attenuated MMP13-mediated BACE1 regulation. In APPswe/PS1E9 mice, an animal model of Alzheimer's disease, hippocampal Mmp13 knockdown or intraperitoneal CL82198 administration reduced BACE1 protein levels and the related amyloid-β precursor protein processing, amyloid-β load and eIF4B phosphorylation, whereas spatial and associative learning and memory performances were improved. Collectively, MMP13 inhibition/CL82198 treatment exhibited therapeutic potential for Alzheimer's disease, via the translational regulation of BACE1.
Figures




Similar articles
-
MiR-340 Reduces the Accumulation of Amyloid-β Through Targeting BACE1 (β-site Amyloid Precursor Protein Cleaving Enzyme 1) in Alzheimer's Disease.Curr Neurovasc Res. 2020;17(1):86-92. doi: 10.2174/1567202617666200117103931. Curr Neurovasc Res. 2020. PMID: 31957613
-
Swedish mutant APP-based BACE1 binding site peptide reduces APP β-cleavage and cerebral Aβ levels in Alzheimer's mice.Sci Rep. 2015 Jun 19;5:11322. doi: 10.1038/srep11322. Sci Rep. 2015. PMID: 26091071 Free PMC article.
-
Mitochondrial TXN2 attenuates amyloidogenesis via selective inhibition of BACE1 expression.J Neurochem. 2021 May;157(4):1351-1365. doi: 10.1111/jnc.15184. Epub 2020 Sep 30. J Neurochem. 2021. PMID: 32920833
-
Inhibition of sphingomyelin synthase 1 ameliorates alzheimer-like pathology in APP/PS1 transgenic mice through promoting lysosomal degradation of BACE1.Exp Neurol. 2019 Jan;311:67-79. doi: 10.1016/j.expneurol.2018.09.012. Epub 2018 Sep 20. Exp Neurol. 2019. PMID: 30243987
-
Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer's disease.Prog Neurobiol. 2006 Jun;79(2):95-111. doi: 10.1016/j.pneurobio.2006.06.001. Epub 2006 Aug 14. Prog Neurobiol. 2006. PMID: 16904810 Review.
Cited by
-
Myeloid ectopic viral integration site 2 accelerates the progression of Alzheimer's disease.Aging Cell. 2024 Oct;23(10):e14260. doi: 10.1111/acel.14260. Epub 2024 Jul 12. Aging Cell. 2024. PMID: 38994634 Free PMC article.
-
Decay in Retinoic Acid Signaling in Varied Models of Alzheimer's Disease and In-Vitro Test of Novel Retinoic Acid Receptor Ligands (RAR-Ms) to Regulate Protective Genes.J Alzheimers Dis. 2020;73(3):935-954. doi: 10.3233/JAD-190931. J Alzheimers Dis. 2020. PMID: 31884477 Free PMC article.
-
Comparative Study of Protein Aggregation Propensity and Mutation Tolerance Between Naked Mole-Rat and Mouse.Genome Biol Evol. 2022 May 3;14(5):evac057. doi: 10.1093/gbe/evac057. Genome Biol Evol. 2022. PMID: 35482036 Free PMC article.
-
SHMT2 Mediates Small-Molecule-Induced Alleviation of Alzheimer Pathology Via the 5'UTR-dependent ADAM10 Translation Initiation.Adv Sci (Weinh). 2024 Mar;11(11):e2305260. doi: 10.1002/advs.202305260. Epub 2024 Jan 6. Adv Sci (Weinh). 2024. PMID: 38183387 Free PMC article.
-
Arc Regulates Transcription of Genes for Plasticity, Excitability and Alzheimer's Disease.Biomedicines. 2022 Aug 11;10(8):1946. doi: 10.3390/biomedicines10081946. Biomedicines. 2022. PMID: 36009494 Free PMC article.
References
-
- Chen JM, Nelson FC, Levin JI, Mobilio D, Moy FJ, Nilakantan R et al. . Structure-based design of a novel, potent, and selective inhibitor for MMP-13 utilizing NMR spectroscopy and computer-aided molecular design. J Am Chem Soc 2000; 122: 9648–54.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

