Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5'UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication
- PMID: 30597844
- PMCID: PMC6357077
- DOI: 10.3390/v11010017
Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5'UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication
Abstract
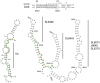
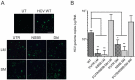
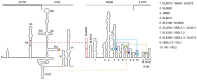
The hepatitis C virus (HCV) genome contains structured elements thought to play important regulatory roles in viral RNA translation and replication processes. We used in vitro RNA binding assays to map interactions involving the HCV 5'UTR and distal sequences in NS5B to examine their impact on viral RNA replication. The data revealed that 5'UTR nucleotides (nt) 95⁻110 in the internal ribosome entry site (IRES) domain IIa and matching nt sequence 8528⁻8543 located in the RNA-dependent RNA polymerase coding region NS5B, form a high-affinity RNA-RNA complex in vitro. This duplex is composed of both wobble and Watson-Crick base-pairings, with the latter shown to be essential to the formation of the high-affinity duplex. HCV genomic RNA constructs containing mutations in domain IIa nt 95⁻110 or within the genomic RNA location comprising nt 8528⁻8543 displayed, on average, 5-fold less intracellular HCV RNA and 6-fold less infectious progeny virus. HCV genomic constructs containing complementary mutations for IRES domain IIa nt 95⁻110 and NS5B nt 8528⁻8543 restored intracellular HCV RNA and progeny virus titers to levels obtained for parental virus RNA. We conclude that this long-range duplex interaction between the IRES domain IIa and NS5B nt 8528⁻8543 is essential for optimal virus replication.
Keywords: Flaviviridae; HCV IRES; NS5B; RNA folding; RNA stem-loop; circular RNA; long-distance RNA–RNA interaction; secondary structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5' untranslated region and directs HCV RNA replication through circularizing the viral genome.J Virol. 2011 Aug;85(16):7954-64. doi: 10.1128/JVI.00339-11. Epub 2011 Jun 1. J Virol. 2011. PMID: 21632751 Free PMC article.
-
Alterations to both the primary and predicted secondary structure of stem-loop IIIc of the hepatitis C virus 1b 5' untranslated region (5'UTR) lead to mutants severely defective in translation which cannot be complemented in trans by the wild-type 5'UTR sequence.J Virol. 1999 Mar;73(3):2359-64. doi: 10.1128/JVI.73.3.2359-2364.1999. J Virol. 1999. PMID: 9971819 Free PMC article.
-
cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5' nontranslated region chimeras.RNA. 1998 Nov;4(11):1418-35. doi: 10.1017/s1355838298981031. RNA. 1998. PMID: 9814762 Free PMC article.
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis.Viruses. 2012 Oct 19;4(10):2233-50. doi: 10.3390/v4102233. Viruses. 2012. PMID: 23202462 Free PMC article. Review.
Cited by
-
Activation of viral transcription by stepwise largescale folding of an RNA virus genome.Nucleic Acids Res. 2020 Sep 18;48(16):9285-9300. doi: 10.1093/nar/gkaa675. Nucleic Acids Res. 2020. PMID: 32785642 Free PMC article.
-
Hepatitis C Viral Replication Complex.Viruses. 2021 Mar 22;13(3):520. doi: 10.3390/v13030520. Viruses. 2021. PMID: 33809897 Free PMC article. Review.
-
The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle.Int J Mol Sci. 2020 Feb 21;21(4):1479. doi: 10.3390/ijms21041479. Int J Mol Sci. 2020. PMID: 32098260 Free PMC article. Review.
References
-
- Lindenbach B.D., Murry C.L., Thiel J.M., Rice C.M., Knipe D.M., Howley P.M. Virology, Fields. Volume 6. Lippincott, Williams and Wilkins; Philadelphia, PA, USA: 2013. Flaviviridae; pp. 712–746.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

