Advanced lipoxidation end products (ALEs) as RAGE binders: Mass spectrometric and computational studies to explain the reasons why
- PMID: 30598328
- PMCID: PMC6859533
- DOI: 10.1016/j.redox.2018.101083
Advanced lipoxidation end products (ALEs) as RAGE binders: Mass spectrometric and computational studies to explain the reasons why
Abstract
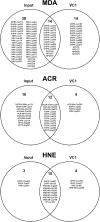
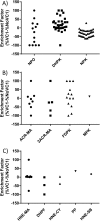
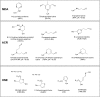
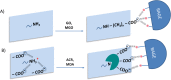
Advanced Lipoxidation End-products (ALEs) are modified proteins that can act as pathogenic factors in several chronic diseases. Several molecular mechanisms have so far been considered to explain the damaging action of ALEs and among these a pathway involving the receptor for advanced glycation end products (RAGE) should be considered. The aim of the present work is to understand if ALEs formed from lipid peroxidation derived reactive carbonyl species (RCS) are able to act as RAGE binders and also to gain a deeper insight into the molecular mechanisms involved in the protein-protein engagement. ALEs were produced in vitro, by incubating human serum albumin (HSA) with 4-hydroxy-trans- 2-nonenal (HNE), acrolein (ACR) and malondialdehyde (MDA). The identification of ALEs was performed by MS. ALEs were then subjected to the VC1 Pull-Down assay (VC1 is the ligand binding domain of RAGE) and the enrichment factor (the difference between the relative abundance in the enriched sample minus the amount in the untreated one) as an index of affinity, was determined. Computation studies were then carried out to explain the factors governing the affinity of the adducted moieties and the site of interaction on adducted HSA for VC1-binding. The in silico analyses revealed the key role played by those adducts which strongly reduce the basicity of the modified residues and thus occur at their neutral state at physiological conditions (e.g. the MDA adducts, dihydropyridine-Lysine (DHPK) and N-2-pyrimidyl-ornithine (NPO), and acrolein derivatives, N-(3-formyl-3,4-dehydro-piperidinyl) lysine, FDPK). These neutral adducts become unable to stabilize ion-pairs with the surrounding negative residues which thus can contact the RAGE positive residues. In conclusion, ALEs derived from lipid peroxidation-RCS are binders of RAGE and this affinity depends on the effect of the adduct moiety to reduce the basicity of the target amino acid and on the acid moieties surrounding the aminoacidic target.
Keywords: 4-hydroxy-trans− 2-nonenal (HNE); Acrolein (ACR) and malondialdehyde (MDA); Advanced lipoxidation end products (ALEs); Human serum albumin (HSA); Pull-down assay; RAGE; Reactive Carbonyl Species (RCS); VC1 domain.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Gueraud F., Atalay M., Bresgen N., Cipak A., Eckl P.M., Huc L., Jouanin I., Siems W., Uchida K. Chemistry and biochemistry of lipid peroxidation products. Free Radic. Res. 2010;44(10):1098–1124. - PubMed
-
- Vistoli G., De Maddis D., Cipak A., Zarkovic N., Carini M., Aldini G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): an overview of their mechanisms of formation. Free Radic. Res. 2013;47(Suppl 1):3–27. - PubMed
-
- Aldini G., Vistoli G., Stefek M., Chondrogianni N., Grune T., Sereikaite J., Sadowska-Bartosz I., Bartosz G. Molecular strategies to prevent, inhibit, and degrade advanced glycoxidation and advanced lipoxidation end products. Free Radic. Res. 2013;47(Suppl 1):93–137. - PubMed
-
- Mol M., Regazzoni L., Altomare A., Degani G., Carini M., Vistoli G., Aldini G. Enzymatic and non-enzymatic detoxification of 4-hydroxynonenal: methodological aspects and biological consequences. Free Radic. Biol. Med. 2017;111:328–344. - PubMed
-
- Curtis T. The role lipid aldehydes and ALEs in the pathogenesis of diabetic retinopathy. Free Radic. Biol. Med. 2014;75(Suppl 1):S8. - PubMed

