RIPK1 and Caspase-8 Ensure Chromosome Stability Independently of Their Role in Cell Death and Inflammation
- PMID: 30598363
- PMCID: PMC6375735
- DOI: 10.1016/j.molcel.2018.11.010
RIPK1 and Caspase-8 Ensure Chromosome Stability Independently of Their Role in Cell Death and Inflammation
Abstract
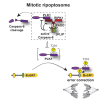
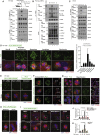
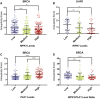
Receptor-interacting protein kinase (RIPK) 1 functions as a key mediator of tissue homeostasis via formation of Caspase-8 activating ripoptosome complexes, positively and negatively regulating apoptosis, necroptosis, and inflammation. Here, we report an unanticipated cell-death- and inflammation-independent function of RIPK1 and Caspase-8, promoting faithful chromosome alignment in mitosis and thereby ensuring genome stability. We find that ripoptosome complexes progressively form as cells enter mitosis, peaking at metaphase and disassembling as cells exit mitosis. Genetic deletion and mitosis-specific inhibition of Ripk1 or Caspase-8 results in chromosome alignment defects independently of MLKL. We found that Polo-like kinase 1 (PLK1) is recruited into mitotic ripoptosomes, where PLK1's activity is controlled via RIPK1-dependent recruitment and Caspase-8-mediated cleavage. A fine balance of ripoptosome assembly is required as deregulated ripoptosome activity modulates PLK1-dependent phosphorylation of downstream effectors, such as BUBR1. Our data suggest that ripoptosome-mediated regulation of PLK1 contributes to faithful chromosome segregation during mitosis.
Keywords: BUBR1; PLK1; RIPK1; cancer; caspase-8; cell cycle; cell death; chromosomal instability; mitosis; ripoptosome.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004;36:744–749. - PubMed
-
- Berger S.B., Kasparcova V., Hoffman S., Swift B., Dare L., Schaeffer M., Capriotti C., Cook M., Finger J., Hughes-Earle A. Cutting Edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 2014;192:5476–5480. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

