Comprehensive molecular and immunological characterization of hepatocellular carcinoma
- PMID: 30598371
- PMCID: PMC6412165
- DOI: 10.1016/j.ebiom.2018.12.058
Comprehensive molecular and immunological characterization of hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is a heterogeneous disease with various etiological factors, and ranks as the second leading cause of cancer-related mortality worldwide due to multi-focal recurrence. We herein identified three major subtypes of HCC by performing integrative analysis of two omics data sets, and clarified that this classification was closely correlated with clinicopathological factors, immune profiles and recurrence patterns.
Methods: In the test study, 183 tumor specimens surgically resected from HCC patients were collected for unsupervised clustering analysis of gene expression signatures and comparative analysis of gene mutations. These results were validated by using genome, methylome and transcriptome data of 373 HCC patients provided from the Cancer Genome Atlas Network. In addition, omics data were obtained from pairs of primary and recurrent HCC.
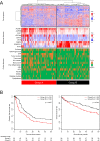
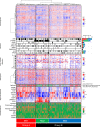
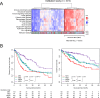
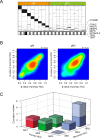
Findings: Comprehensive molecular evaluation of HCC by multi-platform analysis defined three major subtypes: (1) mitogenic and stem cell-like tumors with chromosomal instability; (2) CTNNB1-mutated tumors displaying immune suppression; and (3) metabolic disease-associated tumors, which included an immunogenic subgroup characterized by macrophage infiltration and favorable prognosis. Although genomic and epigenomic analysis explicitly distinguished between HCC with intrahepatic metastasis (IM) and multi-centric HCC (MC), the phenotypic similarity between the primary and recurrent tumors was not correlated to the IM/MC origin, but to the classification.
Interpretation: Identification of these HCC subtypes provides further insights into patient stratification as well as presents opportunities for therapeutic development. FUND: Ministry of Education, Culture, Sports, Science and Technology of Japan (16H02670 and 18K19575), Japan Agency for Medical Research and Development (JP15cm0106064, JP17cm0106518, JP18cm0106540 and JP18fk0210040).
Keywords: CTNNB1 mutation; Hepatocellular carcinoma; Hepatocellular carcinoma with intrahepatic metastasis; Immunogenic cancer; Integrative analysis; Metabolic disease associated cancer; Molecular classification; Multi-centric hepatocellular carcinoma.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. - PubMed
-
- de Bono J.S., Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467(7315):543–549. - PubMed
-
- Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–764. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

