First Report of Chlamydia abortus in Farmed Fur Animals
- PMID: 30598995
- PMCID: PMC6287152
- DOI: 10.1155/2018/4289648
First Report of Chlamydia abortus in Farmed Fur Animals
Abstract
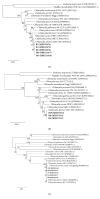
Chlamydia (C.) abortus, a globally distributed obligate intracellular bacterium, has attracted increasing interest according to its veterinary importance and zoonotic nature. C. abortus can infect a variety of animals and cause foetal loss in livestock resulting in economic loss. In this study, the samples collected from two farms of foxes (n=20), raccoon dogs (n=15) and minks (n=20), were investigated by Chlamydiaceae- and Chlamydia species-specific real-time PCR. The results showed that all the tested foxes (20/20) and raccoon dogs (15/15) harbored Chlamydia spp., while 5% of minks (1/20) were positive for Chlamydia spp. C. abortus was identified in all positive samples as the dominant Chlamydia species, with C. pecorum DNA coexistence in some of the rectal samples (7/20) taken from foxes. Phylogenetic analysis based on specific gene fragments of 16S rRNA, IGS-23S rRNA, and ompA revealed that all sequences obtained in this study were assigned to the Chlamydiaceae family with high similarity to C. abortus S26/3 and B577 previously identified in ruminants. This is the first report confirming that farmed foxes, raccoon dogs, and minks carry C. abortus. Further studies are needed to fully elucidate the epidemiology and pathogenicity of this pathogen in farmed fur animals as well as the potential risks to public health.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

