Celastrol-loaded PEG-b-PPS nanocarriers as an anti-inflammatory treatment for atherosclerosis
- PMID: 30601470
- PMCID: PMC6368678
- DOI: 10.1039/c8bm01224e
Celastrol-loaded PEG-b-PPS nanocarriers as an anti-inflammatory treatment for atherosclerosis
Abstract
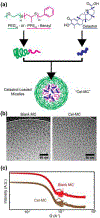
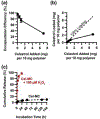
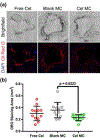
In this work, the hydrophobic small molecule NF-κB inhibitor celastrol was loaded into poly(ethylene glycol)-b-poly(propylene sulfide) (PEG-b-PPS) micelles. PEG-b-PPS micelles demonstrated high loading efficiency, low polydispersity, and no morphological changes upon loading with celastrol. Encapsulation of celastrol within these nanocarriers significantly reduced cytotoxicity compared to free celastrol, while simultaneously expanding the lower concentration range for effective inhibition of NF-κB signaling by nearly 50 000-fold. Furthermore, celastrol-loaded micelles successfully reduced TNF-α secretion after LPS stimulation of RAW 264.7 cells and reduced the number of neutrophils and inflammatory monocytes within atherosclerotic plaques of ldlr-/- mice. This reduction in inflammatory cells was matched by a reduction in plaque area, suggesting that celastrol-loaded nanocarriers may serve as an anti-inflammatory treatment for atherosclerosis.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare.
Figures



Similar articles
-
Inhibition of NF-kappa B activation through targeting I kappa B kinase by celastrol, a quinone methide triterpenoid.Biochem Pharmacol. 2006 Nov 15;72(10):1311-21. doi: 10.1016/j.bcp.2006.08.014. Epub 2006 Sep 18. Biochem Pharmacol. 2006. PMID: 16984800
-
Modulation of Schlemm's canal endothelial cell stiffness via latrunculin loaded block copolymer micelles.J Biomed Mater Res A. 2018 Jul;106(7):1771-1779. doi: 10.1002/jbm.a.36376. Epub 2018 Mar 6. J Biomed Mater Res A. 2018. PMID: 29468812 Free PMC article.
-
Celastrol nanoparticles inhibit corneal neovascularization induced by suturing in rats.Int J Nanomedicine. 2012;7:1163-73. doi: 10.2147/IJN.S27860. Epub 2012 Mar 1. Int J Nanomedicine. 2012. PMID: 22419865 Free PMC article.
-
Celastrol, a potent antioxidant and anti-inflammatory drug, as a possible treatment for Alzheimer's disease.Prog Neuropsychopharmacol Biol Psychiatry. 2001 Oct;25(7):1341-57. doi: 10.1016/s0278-5846(01)00192-0. Prog Neuropsychopharmacol Biol Psychiatry. 2001. PMID: 11513350 Review.
-
Celastrol: Molecular targets of Thunder God Vine.Biochem Biophys Res Commun. 2010 Apr 9;394(3):439-42. doi: 10.1016/j.bbrc.2010.03.050. Epub 2010 Mar 10. Biochem Biophys Res Commun. 2010. PMID: 20226165 Review.
Cited by
-
Inflammation-responsive delivery systems for the treatment of chronic inflammatory diseases.Drug Deliv Transl Res. 2021 Aug;11(4):1475-1497. doi: 10.1007/s13346-021-00977-8. Epub 2021 Apr 15. Drug Deliv Transl Res. 2021. PMID: 33860447 Free PMC article. Review.
-
Sustained release of drug-loaded nanoparticles from injectable hydrogels enables long-term control of macrophage phenotype.Biomater Sci. 2022 Dec 6;10(24):6951-6967. doi: 10.1039/d2bm01113a. Biomater Sci. 2022. PMID: 36341688 Free PMC article.
-
Targeting Early Atherosclerosis: A Focus on Oxidative Stress and Inflammation.Oxid Med Cell Longev. 2019 Jul 1;2019:8563845. doi: 10.1155/2019/8563845. eCollection 2019. Oxid Med Cell Longev. 2019. PMID: 31354915 Free PMC article. Review.
-
Bioresponsive drug delivery systems for the treatment of inflammatory diseases.J Control Release. 2020 Nov 10;327:641-666. doi: 10.1016/j.jconrel.2020.09.008. Epub 2020 Sep 8. J Control Release. 2020. PMID: 32911014 Free PMC article. Review.
-
Celastrol: A lead compound that inhibits SARS-CoV-2 replication, the activity of viral and human cysteine proteases, and virus-induced IL-6 secretion.Drug Dev Res. 2022 Nov;83(7):1623-1640. doi: 10.1002/ddr.21982. Epub 2022 Aug 21. Drug Dev Res. 2022. PMID: 35989498 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

