Roles of Oncogenic Long Non-coding RNAs in Cancer Development
- PMID: 30602079
- PMCID: PMC6440676
- DOI: 10.5808/GI.2018.16.4.e18
Roles of Oncogenic Long Non-coding RNAs in Cancer Development
Abstract
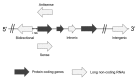
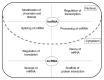
Long non-coding RNAs (lncRNAs) are classified as RNAs that are longer than 200 nucleotides and cannot be translated into protein. Several studies have demonstrated that lncRNAs are directly or indirectly involved in a variety of biological processes and in the regulation of gene expression. In addition, lncRNAs have important roles in many diseases including cancer. It has been shown that abnormal expression of lncRNAs is observed in several human solid tumors. Several studies have shown that many lncRNAs can function as oncogenes in cancer development through the induction of cell cycle progression, cell proliferation and invasion, anti-apoptosis, and metastasis. Oncogenic lncRNAs have the potential to become promising biomarkers and might be potent prognostic targets in cancer therapy. However, the biological and molecular mechanisms of lncRNA involvement in tumorigenesis have not yet been fully elucidated. This review summarizes studies on the regulatory and functional roles of oncogenic lncRNAs in the development and progression of various types of cancer.
Keywords: breast cancer; colorectal cancer; glioblastoma; long non-coding RNA; non-small cell lung cancer; oncogenes.
Conflict of interest statement
No potential conflicts of interest relevant to this article was reported.
Figures


Similar articles
-
Tumor suppressor and oncogenic role of long non-coding RNAs in cancer.North Clin Istanb. 2019 Nov 22;7(1):81-86. doi: 10.14744/nci.2019.46873. eCollection 2020. North Clin Istanb. 2019. PMID: 32232211 Free PMC article. Review.
-
The role of long non-coding RNA AFAP1-AS1 in human malignant tumors.Pathol Res Pract. 2018 Oct;214(10):1524-1531. doi: 10.1016/j.prp.2018.08.014. Epub 2018 Aug 20. Pathol Res Pract. 2018. PMID: 30173945 Review.
-
Roles of Wnt/β-Catenin Signaling Pathway Regulatory Long Non-Coding RNAs in the Pathogenesis of Non-Small Cell Lung Cancer.Cancer Manag Res. 2020 Jun 3;12:4181-4191. doi: 10.2147/CMAR.S241519. eCollection 2020. Cancer Manag Res. 2020. PMID: 32581590 Free PMC article. Review.
-
Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy.Med Oncol. 2016 Feb;33(2):18. doi: 10.1007/s12032-016-0731-2. Epub 2016 Jan 19. Med Oncol. 2016. PMID: 26786153 Review.
-
The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring.Crit Rev Clin Lab Sci. 2014 Dec;51(6):344-57. doi: 10.3109/10408363.2014.944299. Epub 2014 Aug 15. Crit Rev Clin Lab Sci. 2014. PMID: 25123609 Review.
Cited by
-
Gene Expression Profiling Regulated by lncRNA H19 Using Bioinformatic Analyses in Glioma Cell Lines.Cancer Genomics Proteomics. 2024 Nov-Dec;21(6):608-621. doi: 10.21873/cgp.20477. Cancer Genomics Proteomics. 2024. PMID: 39467632 Free PMC article.
-
Association between genetic polymorphisms of long noncoding RNA H19 and cancer risk: a meta-analysis.J Genet. 2019 Sep;98:81. J Genet. 2019. PMID: 31544800 Review.
-
Expression and Alteration Value of Long Noncoding RNA AB073614 and FER1L4 in Patients with Acute Myeloid Leukemia (AML).Asian Pac J Cancer Prev. 2023 Jul 1;24(7):2271-2277. doi: 10.31557/APJCP.2023.24.7.2271. Asian Pac J Cancer Prev. 2023. PMID: 37505756 Free PMC article.
-
Downregulation of long non‑coding RNA GACAT1 suppresses proliferation and induces apoptosis of NSCLC cells by sponging microRNA‑422a.Int J Mol Med. 2021 Feb;47(2):659-667. doi: 10.3892/ijmm.2020.4826. Epub 2020 Dec 22. Int J Mol Med. 2021. Retraction in: Int J Mol Med. 2024 Jan;53(1):1. doi: 10.3892/ijmm.2023.5325. PMID: 33416153 Free PMC article. Retracted.
-
Role of Non-Coding RNAs in Colorectal Cancer: Focus on Long Non-Coding RNAs.Int J Mol Sci. 2022 Nov 3;23(21):13431. doi: 10.3390/ijms232113431. Int J Mol Sci. 2022. PMID: 36362222 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
