PRAME promotes epithelial-to-mesenchymal transition in triple negative breast cancer
- PMID: 30602372
- PMCID: PMC6317205
- DOI: 10.1186/s12967-018-1757-3
PRAME promotes epithelial-to-mesenchymal transition in triple negative breast cancer
Abstract
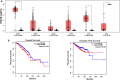
Background: The triple negative breast cancer (TNBC) paradox marks a major challenge in the treatment-decision making process. TNBC patients generally respond better to neoadjuvant chemotherapy compared to other breast cancer patients; however, they have a substantial higher risk of disease recurrence. We evaluated the expression of the tumor-associated antigen PReferentially Antigen expressed in MElanoma (PRAME) as a prognostic biomarker in breast cancer and explored its role in cell migration and invasion, key hallmarks of progressive and metastatic disease.
Methods: TCGA and GTeX datasets were interrogated to assess the expression of PRAME in relation to overall and disease-free survival. The role of PRAME in cell migration and invasion was investigated using gain- and loss-of-function TNBC cell line models.
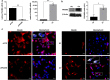
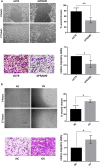
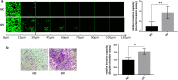
Results: We show that PRAME promotes migration and invasion of TNBC cells through changes in expression of E-cadherin, N-cadherin, vimentin and ZEB1, core markers of an epithelial-to-mesenchymal transition. Mechanistic analysis of PRAME-overexpressing cells showed an upregulation of 11 genes (SNAI1, TCF4, TWIST1, FOXC2, IL1RN, MMP2, SOX10, WNT11, MMP3, PDGFRB, and JAG1) and downregulation of 2 genes (BMP7 and TSPAN13). Gene ontology analyses revealed enrichment of genes that are dysregulated in ovarian and esophageal cancer and are involved in transcription and apoptosis. In line with this, interrogation of TCGA and GTEx data demonstrated an increased PRAME expression in ovarian and esophageal tumor tissues in addition to breast tumors where it is associated with worse survival.
Conclusions: Our findings indicate that PRAME plays a tumor-promoting role in triple negative breast cancer by increasing cancer cell motility through EMT-gene reprogramming. Therefore, PRAME could serve as a prognostic biomarker and/or therapeutic target in TNBC.
Keywords: Epithelial-to-mesenchymal transition; Invasion; Migration; PRAME; PReferentially Antigen expressed in Melanoma; Triple negative breast cancer.
Figures



References
-
- World Health Organization . World Cancer Report 2014; International Agency for Research on Cancers. Lyon: IARC Press; 2003.
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Van De Rijn M, Jeffrey SS, Thorsen T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

