The Transcriptional Cycle Is Suited to Daytime N2 Fixation in the Unicellular Cyanobacterium " Candidatus Atelocyanobacterium thalassa" (UCYN-A)
- PMID: 30602582
- PMCID: PMC6315102
- DOI: 10.1128/mBio.02495-18
The Transcriptional Cycle Is Suited to Daytime N2 Fixation in the Unicellular Cyanobacterium " Candidatus Atelocyanobacterium thalassa" (UCYN-A)
Abstract
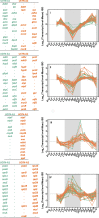
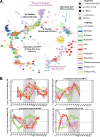
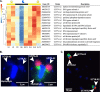
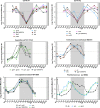
Symbiosis between a marine alga and a N2-fixing cyanobacterium (Cyanobacterium UCYN-A) is geographically widespread in the oceans and is important in the marine N cycle. UCYN-A is uncultivated and is an unusual unicellular cyanobacterium because it lacks many metabolic functions, including oxygenic photosynthesis and carbon fixation, which are typical in cyanobacteria. It is now presumed to be an obligate symbiont of haptophytes closely related to Braarudosphaera bigelowii N2-fixing cyanobacteria use different strategies to avoid inhibition of N2 fixation by the oxygen evolved in photosynthesis. Most unicellular cyanobacteria temporally separate the two incompatible activities by fixing N2 only at night, but, surprisingly, UCYN-A appears to fix N2 during the day. The goal of this study was to determine how the unicellular UCYN-A strain coordinates N2 fixation and general metabolism compared to other marine cyanobacteria. We found that UCYN-A has distinct daily cycles of many genes despite the fact that it lacks two of the three circadian clock genes found in most cyanobacteria. We also found that the transcription patterns in UCYN-A are more similar to those in marine cyanobacteria that are capable of aerobic N2 fixation in the light, such as Trichodesmium and heterocyst-forming cyanobacteria, than to those in Crocosphaera or Cyanothece species, which are more closely related to unicellular marine cyanobacteria evolutionarily. Our findings suggest that the symbiotic interaction has resulted in a shift of transcriptional regulation to coordinate UCYN-A metabolism with that of the phototrophic eukaryotic host, thus allowing efficient coupling of N2 fixation (by the cyanobacterium) to the energy obtained from photosynthesis (by the eukaryotic unicellular alga) in the light.IMPORTANCE The symbiotic N2-fixing cyanobacterium UCYN-A, which is closely related to Braarudosphaera bigelowii, and its eukaryotic algal host have been shown to be globally distributed and important in open-ocean N2 fixation. These unique cyanobacteria have reduced metabolic capabilities, even lacking genes for oxygenic photosynthesis and carbon fixation. Cyanobacteria generally use energy from photosynthesis for nitrogen fixation but require mechanisms for avoiding inactivation of the oxygen-sensitive nitrogenase enzyme by ambient oxygen (O2) or the O2 evolved through photosynthesis. This study showed that symbiosis between the N2-fixing cyanobacterium UCYN-A and its eukaryotic algal host has led to adaptation of its daily gene expression pattern in order to enable daytime aerobic N2 fixation, which is likely more energetically efficient than fixing N2 at night, as found in other unicellular marine cyanobacteria.
Keywords: cyanobacteria; diel cycle; marine microbiology; nitrogen fixation; symbiosis; whole-genome expression.
Copyright © 2019 Muñoz-Marín et al.
Figures


References
-
- Postgate JR. 1982. The fundamentals of nitrogen fixation. Cambridge University Press, London, United Kingdom.
-
- Martínez-Pérez C, Mohr W, Löscher C, Dekaezemacker J, Littmann S, Yilmaz P, Lehnen N, Fuchs BM, Lavik G, Schmitz R, LaRoche J, Kuypers MM. 2016. The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat Microbiol 1:16163. doi: 10.1038/NMICROBIOL.2016.163. - DOI - PubMed
-
- Farnelid H, Turk-Kubo K, Muñoz-Marín MC, Zehr JP. 2016. New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquat Microb Ecol 77:125–138. doi: 10.3354/ame01794. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

