Lund Human Mesencephalic (LUHMES) Neuronal Cell Line Supports Herpes Simplex Virus 1 Latency In Vitro
- PMID: 30602607
- PMCID: PMC6401467
- DOI: 10.1128/JVI.02210-18
Lund Human Mesencephalic (LUHMES) Neuronal Cell Line Supports Herpes Simplex Virus 1 Latency In Vitro
Abstract
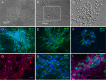
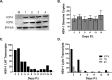
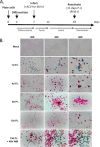
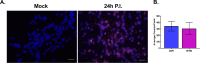
Lund human mesencephalic (LUHMES) cells are human embryonic neuronal precursor cells that can be maintained as proliferating cells due to the expression of a tetracycline-regulatable (Tet-off) v-myc transgene. They can be differentiated to postmitotic neurons by the addition of tetracycline, glial cell-derived neurotrophic factor (GDNF), and dibutyryl cAMP. We demonstrate that these cells can be infected with herpes simplex virus 1 (HSV-1) at a multiplicity of infection (MOI) of 3 with the majority of cells surviving. By 6 days postinfection, there is a loss of lytic gene transcription and an increase in the numbers of neurons that express the latency-associated transcripts (LATs). Importantly, the virus can then be reactivated by the addition of a phosphoinositide 3-kinase inhibitor, which has previously been shown to reactivate HSV-1 in rat neuron cultures. While rodent primary culture neuron systems have been described, these are limited by their lack of scalability, as it is difficult to obtain more than 500,000 neurons to employ for a given experiment. Several recent papers have described a human dorsal root ganglion (DRG) neuron culture model and human induced pleuripotent stem cell (iPSC) neuron culture models that are scalable, but they require that the presence of an antiviral suppression be maintained following HSV-1 infection. The human LUHMES cell model of HSV-1 infection described here may be especially useful for studying HSV-1 latency and reactivation on account of its scalability, its amenability to maintenance of latency without the continual use of antiviral inhibitors, and its latent gene expression profile which mirrors many properties observed in vivo, importantly, the heterogeneity of cells expressing the LATs.IMPORTANCE Herpes simplex virus (HSV) is responsible for significant morbidity in humans due to its ability to cause oral and genital lesions, ocular disease, and encephalitis. While antivirals can attenuate the severity and frequency of disease, there is no vaccine or cure. Understanding the molecular details of HSV latency and reactivation is key to the development of new therapies. One of the difficulties in studying HSV latency has been the need to rely on establishment of latent infections in animal models. While rodent primary neuron culture models have shown promise, they yield relatively small numbers of latently infected neurons for biochemical and molecular analyses. Here we present the use of a human central nervous system (CNS)-derived conditionally proliferating cell line that can be differentiated into mature neurons and latently infected with HSV-1. This model shows promise as a scalable tool to study molecular and biochemical aspects of HSV-1 latency and reactivation in human neurons.
Keywords: latency; neuron.
Copyright © 2019 American Society for Microbiology.
Figures

Similar articles
-
Herpes Simplex Virus 1 MicroRNA miR-H8 Is Dispensable for Latency and Reactivation In Vivo.J Virol. 2021 Jan 28;95(4):e02179-20. doi: 10.1128/JVI.02179-20. Print 2021 Jan 28. J Virol. 2021. PMID: 33208453 Free PMC article.
-
Modulation of Voltage-Gated Sodium Channel Activity in Human Dorsal Root Ganglion Neurons by Herpesvirus Quiescent Infection.J Virol. 2020 Jan 17;94(3):e01823-19. doi: 10.1128/JVI.01823-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31694955 Free PMC article.
-
An Immortalized Human Dorsal Root Ganglion Cell Line Provides a Novel Context To Study Herpes Simplex Virus 1 Latency and Reactivation.J Virol. 2017 May 26;91(12):e00080-17. doi: 10.1128/JVI.00080-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28404842 Free PMC article.
-
A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review.
-
Molecular circuitry regulating herpes simplex virus type 1 latency in neurons.J Neurovirol. 2000 Feb;6(1):6-24. doi: 10.3109/13550280009006378. J Neurovirol. 2000. PMID: 10786993 Review.
Cited by
-
Transcriptional Regulation of Latency-Associated Transcripts (LATs) of Herpes Simplex Viruses.J Cancer. 2020 Mar 5;11(11):3387-3399. doi: 10.7150/jca.40186. eCollection 2020. J Cancer. 2020. PMID: 32231745 Free PMC article.
-
Current Approaches and Tools Used in Drug Development against Parkinson's Disease.Biomolecules. 2021 Jun 16;11(6):897. doi: 10.3390/biom11060897. Biomolecules. 2021. PMID: 34208760 Free PMC article. Review.
-
A viral lncRNA tethers HSV-1 genomes at the nuclear periphery to establish viral latency.J Virol. 2023 Dec 21;97(12):e0143823. doi: 10.1128/jvi.01438-23. Epub 2023 Nov 22. J Virol. 2023. PMID: 37991364 Free PMC article.
-
De Novo Polycomb Recruitment: Lessons from Latent Herpesviruses.Viruses. 2021 Jul 27;13(8):1470. doi: 10.3390/v13081470. Viruses. 2021. PMID: 34452335 Free PMC article. Review.
-
The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection.Virus Res. 2020 Aug;285:198015. doi: 10.1016/j.virusres.2020.198015. Epub 2020 May 13. Virus Res. 2020. PMID: 32416261 Free PMC article. Review.
References
-
- Preston CM, Russell J, Harris RA, Jamieson DR. 1994. Herpes simplex virus latency in tissue culture cells. Gene Ther 1:S49–S50. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

