High Permissiveness for Genetic Exchanges between Enteroviruses of Species A, including Enterovirus 71, Favors Evolution through Intertypic Recombination in Madagascar
- PMID: 30602612
- PMCID: PMC6401452
- DOI: 10.1128/JVI.01667-18
High Permissiveness for Genetic Exchanges between Enteroviruses of Species A, including Enterovirus 71, Favors Evolution through Intertypic Recombination in Madagascar
Abstract
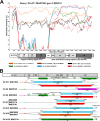
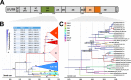
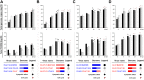
Human enteroviruses of species A (EV-A) are the leading cause of hand-foot-and-mouth disease (HFMD). EV-A71 is frequently implicated in HFMD outbreaks and can also cause severe neurological manifestations. We investigated the molecular epidemiological processes at work and the contribution of genetic recombination to the evolutionary history of EV-A in Madagascar, focusing on the recently described EV-A71 genogroup F in particular. Twenty-three EV-A isolates, collected mostly in 2011 from healthy children living in various districts of Madagascar, were characterized by whole-genome sequencing. Eight different types were identified, highlighting the local circulation and diversity of EV-A. Comparative genome analysis revealed evidence of frequent recent intra- and intertypic genetic exchanges between the noncapsid sequences of Madagascan EV-A isolates. The three EV-A71 isolates had different evolutionary histories in terms of recombination, with one isolate displaying a mosaic genome resulting from recent genetic exchanges with Madagascan coxsackieviruses A7 and possibly A5 and A10 or common ancestors. The engineering and characterization of recombinants generated from progenitors belonging to different EV-A types or EV-A71 genogroups with distantly related nonstructural sequences indicated a high level of permissiveness for intertypic genetic exchange in EV-A. This permissiveness suggests that the primary viral functions associated with the nonstructural sequences have been highly conserved through the diversification and evolution of the EV-A species. No outbreak of disease due to EV-A has yet been reported in Madagascar, but the diversity, circulation, and evolution of these viruses justify surveillance of EV-A circulation and HFMD cases to prevent possible outbreaks due to emerging strains.IMPORTANCE Human enteroviruses of species A (EV-A), including EV-A71, are the leading cause of hand-foot-and-mouth disease (HFMD) and may also cause severe neurological manifestations. We investigated the circulation and molecular evolution of EV-A in Madagascar, focusing particularly on the recently described EV-A71 genogroup F. Eight different types, collected mostly in 2011, were identified, highlighting the local circulation and diversity of EV-A. Comparative genome analysis revealed evidence of frequent genetic exchanges between the different types of isolates. The three EV-A71 isolates had different evolutionary histories in terms of recombination. The engineering and characterization of recombinants involving progenitors belonging to different EV-A types indicated a high degree of permissiveness for genetic exchange in EV-A. No outbreak of disease due to EV-A has yet been reported in Madagascar, but the diversity, circulation, and evolution of these viruses justify the surveillance of EV-A circulation to prevent possible HFMD outbreaks due to emerging strains.
Keywords: RNA virus; emergence; enterovirus; picornavirus; recombination; viral evolution.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
Genetic diversity and evolution of enterovirus A71 subgenogroup C1 from children with hand, foot, and mouth disease in Thailand.Arch Virol. 2021 Aug;166(8):2209-2216. doi: 10.1007/s00705-021-05130-x. Epub 2021 Jun 4. Arch Virol. 2021. PMID: 34086143
-
Discovery of Enterovirus A71-like nonstructural genomes in recent circulating viruses of the Enterovirus A species.Emerg Microbes Infect. 2018 Jun 21;7(1):111. doi: 10.1038/s41426-018-0107-0. Emerg Microbes Infect. 2018. PMID: 29930332 Free PMC article.
-
Molecular epidemiology and recombination of Enterovirus A71 in mainland China from 1987 to 2017.Int Microbiol. 2021 Aug;24(3):291-299. doi: 10.1007/s10123-021-00164-2. Epub 2021 Feb 19. Int Microbiol. 2021. PMID: 33608776 Free PMC article.
-
Changes in the EV-A71 Genome through Recombination and Spontaneous Mutations: Impact on Virulence.Viruses. 2018 Jun 12;10(6):320. doi: 10.3390/v10060320. Viruses. 2018. PMID: 29895721 Free PMC article. Review.
-
Enterovirus A71: virulence, antigenicity, and genetic evolution over the years.J Biomed Sci. 2019 Oct 21;26(1):81. doi: 10.1186/s12929-019-0574-1. J Biomed Sci. 2019. PMID: 31630680 Free PMC article. Review.
Cited by
-
Recombination in Enteroviruses, a Multi-Step Modular Evolutionary Process.Viruses. 2019 Sep 14;11(9):859. doi: 10.3390/v11090859. Viruses. 2019. PMID: 31540135 Free PMC article. Review.
-
Global Spread of the B5 Subgenotype EV-A71 and the Phylogeographical Analysis of Chinese Migration Events.Front Cell Infect Microbiol. 2020 Sep 25;10:475. doi: 10.3389/fcimb.2020.00475. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33102246 Free PMC article.
-
Human Antibodies to VP4 Inhibit Replication of Enteroviruses Across Subgenotypes and Serotypes, and Enhance Host Innate Immunity.Front Microbiol. 2020 Sep 25;11:562768. doi: 10.3389/fmicb.2020.562768. eCollection 2020. Front Microbiol. 2020. PMID: 33101238 Free PMC article.
-
The Establishment of Infectious Clone and Single Round Infectious Particles for Coxsackievirus A10.Virol Sin. 2020 Aug;35(4):426-435. doi: 10.1007/s12250-020-00198-2. Epub 2020 Mar 6. Virol Sin. 2020. PMID: 32144688 Free PMC article.
-
Genetic diversity and evolution of enterovirus A71 subgenogroup C1 from children with hand, foot, and mouth disease in Thailand.Arch Virol. 2021 Aug;166(8):2209-2216. doi: 10.1007/s00705-021-05130-x. Epub 2021 Jun 4. Arch Virol. 2021. PMID: 34086143
References
-
- Mirand A, Henquell C, Archimbaud C, Ughetto S, Antona D, Bailly JL, Peigue-Lafeuille H. 2012. Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect 18:E110–E118. doi:10.1111/j.1469-0691.2012.03789.x. - DOI - PubMed
-
- Pallansch M, Oberste M, Whitton J. 2013. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p 839–893. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Rancaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

