Modulation of Bacterial sRNAs Activity by Epigenetic Modifications: Inputs from the Eukaryotic miRNAs
- PMID: 30602712
- PMCID: PMC6356536
- DOI: 10.3390/genes10010022
Modulation of Bacterial sRNAs Activity by Epigenetic Modifications: Inputs from the Eukaryotic miRNAs
Abstract
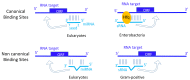
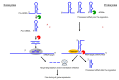
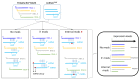
Trans-encoded bacterial regulatory RNAs (sRNAs) are functional analogues of eukaryotic microRNAs (miRNAs). These RNA classes act by base-pairing complementarity with their RNA targets to modulate gene expression (transcription, half-life and/or translation). Based on base-pairing, algorithms predict binding and the impact of small RNAs on targeted-RNAs expression and fate. However, other actors are involved such as RNA binding proteins and epigenetic modifications of the targeted and small RNAs. Post-transcriptional base modifications are widespread in all living organisms where they lower undesired RNA folds through conformation adjustments and influence RNA pairing and stability, especially if remodeling their ends. In bacteria, sRNAs possess RNA modifications either internally (methylation, pseudouridinylation) or at their ends. Nicotinamide adenine dinucleotide were detected at 5'-ends, and polyadenylation can occur at 3'-ends. Eukaryotic miRNAs possess N⁶-methyladenosine (m⁶A), A editing into I, and non-templated addition of uridines at their 3'-ends. Biological functions and enzymes involved in those sRNA and micro RNA epigenetic modifications, when known, are presented and challenged.
Keywords: RNA modifications; bacterial regulatory RNAs; biogenesis; editing; epigenetics; methylation; miRNAs; trans-acting RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Small RNA-mediated regulation in bacteria: A growing palette of diverse mechanisms.Gene. 2018 May 20;656:60-72. doi: 10.1016/j.gene.2018.02.068. Epub 2018 Mar 1. Gene. 2018. PMID: 29501814 Review.
-
Unveiling the orchestration: mycobacterial small RNAs as key mediators in host-pathogen interactions.Front Microbiol. 2024 Jun 5;15:1399280. doi: 10.3389/fmicb.2024.1399280. eCollection 2024. Front Microbiol. 2024. PMID: 38903780 Free PMC article. Review.
-
The Phosphorolytic Exoribonucleases Polynucleotide Phosphorylase and RNase PH Stabilize sRNAs and Facilitate Regulation of Their mRNA Targets.J Bacteriol. 2016 Nov 18;198(24):3309-3317. doi: 10.1128/JB.00624-16. Print 2016 Dec 15. J Bacteriol. 2016. PMID: 27698082 Free PMC article.
-
The Complexity of Posttranscriptional Small RNA Regulatory Networks Revealed by In Silico Analysis of Gossypium arboreum L. Leaf, Flower and Boll Small Regulatory RNAs.PLoS One. 2015 Jun 12;10(6):e0127468. doi: 10.1371/journal.pone.0127468. eCollection 2015. PLoS One. 2015. PMID: 26070200 Free PMC article.
-
A Modular Genetic System for High-Throughput Profiling and Engineering of Multi-Target Small RNAs.Methods Mol Biol. 2018;1737:373-391. doi: 10.1007/978-1-4939-7634-8_21. Methods Mol Biol. 2018. PMID: 29484604
Cited by
-
The Acinetobacter baumannii model can explain the role of small non-coding RNAs as potential mediators of host-pathogen interactions.Front Mol Biosci. 2022 Dec 21;9:1088783. doi: 10.3389/fmolb.2022.1088783. eCollection 2022. Front Mol Biosci. 2022. PMID: 36619166 Free PMC article. Review.
-
Association of oral dysbiosis with oral cancer development.Oncol Lett. 2020 Apr;19(4):3045-3058. doi: 10.3892/ol.2020.11441. Epub 2020 Mar 3. Oncol Lett. 2020. PMID: 32211076 Free PMC article. Review.
-
Interspecies RNA Interactome of Pathogen and Host in a Heritable Defensive Strategy.Front Microbiol. 2021 Jul 21;12:649858. doi: 10.3389/fmicb.2021.649858. eCollection 2021. Front Microbiol. 2021. PMID: 34367078 Free PMC article.
-
In silico identification of human microRNAs pointing centrin genes in Leishmania donovani: Considering the RNAi-mediated gene control.Front Genet. 2024 Feb 8;14:1329339. doi: 10.3389/fgene.2023.1329339. eCollection 2023. Front Genet. 2024. PMID: 38390455 Free PMC article.
-
Benchmarking of five NGS mapping tools for the reference alignment of bacterial outer membrane vesicles-associated small RNAs.Front Microbiol. 2024 Jul 19;15:1401985. doi: 10.3389/fmicb.2024.1401985. eCollection 2024. Front Microbiol. 2024. PMID: 39101033 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

