SG formation relies on eIF4GI-G3BP interaction which is targeted by picornavirus stress antagonists
- PMID: 30603102
- PMCID: PMC6312541
- DOI: 10.1038/s41421-018-0068-4
SG formation relies on eIF4GI-G3BP interaction which is targeted by picornavirus stress antagonists
Abstract
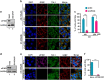
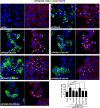
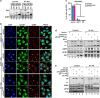
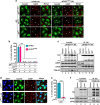
Typical stress granules (tSGs) are stalled translation pre-initiation complex aggregations in the cytoplasm, and their formation is a common consequence of translation initiation inhibition under stress. We previously found that 2A protease of picornaviruses blocks tSG formation and induces atypical SG formation, but the molecular mechanism by which 2A inhibits tSG formation remains unclear. Here, we found that eukaryotic translation initiation factor 4 gamma1 (eIF4GI) is critical for tSG formation by interacting with Ras-GTPase-activating protein SH3-domain-binding protein (G3BP), and this interaction is mediated by aa 182-203 of eIF4GI and the RNA-binding domain of G3BP. Upon eIF4GI-G3BP interaction, eIF4GI can assemble into tSGs and rescue tSG formation. Finally, we found that 2A or L protein of picornaviruses blocks tSG formation by disrupting eIF4GI-G3BP interaction. Our findings provide the first evidence that eIF4GI-G3BP interaction is indispensable for tSG formation, and 2A or L protein of picornaviruses interferes eIF4GI-G3BP interaction, thereby blocking tSG formation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Picornavirus 2A protease regulates stress granule formation to facilitate viral translation.PLoS Pathog. 2018 Feb 7;14(2):e1006901. doi: 10.1371/journal.ppat.1006901. eCollection 2018 Feb. PLoS Pathog. 2018. PMID: 29415027 Free PMC article.
-
G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits.J Cell Biol. 2016 Mar 28;212(7):845-60. doi: 10.1083/jcb.201508028. J Cell Biol. 2016. PMID: 27022092 Free PMC article.
-
Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs.Mol Cell Biol. 2007 Mar;27(6):2324-42. doi: 10.1128/MCB.02300-06. Epub 2007 Jan 8. Mol Cell Biol. 2007. PMID: 17210633 Free PMC article.
-
Translation driven by picornavirus IRES is hampered from Sindbis virus replicons: rescue by poliovirus 2A protease.J Mol Biol. 2010 Sep 10;402(1):101-17. doi: 10.1016/j.jmb.2010.07.014. Epub 2010 Jul 17. J Mol Biol. 2010. PMID: 20643140
-
[G3BP: a promising target for cancer therapy].Yao Xue Xue Bao. 2010 Aug;45(8):945-51. Yao Xue Xue Bao. 2010. PMID: 21351580 Review. Chinese.
Cited by
-
Epigenetic Reprogramming of the Glucose Metabolic Pathways by the Chromatin Effectors During Cancer.Subcell Biochem. 2022;100:269-336. doi: 10.1007/978-3-031-07634-3_9. Subcell Biochem. 2022. PMID: 36301498
-
Recent trends in the elucidation of complex triterpene biosynthetic pathways in horticultural trees.Hortic Res. 2024 Oct 8;12(1):uhae254. doi: 10.1093/hr/uhae254. eCollection 2025 Jan. Hortic Res. 2024. PMID: 39802733 Free PMC article.
-
Porcine Epidemic Diarrhea Virus: An Updated Overview of Virus Epidemiology, Virulence Variation Patterns and Virus-Host Interactions.Viruses. 2022 Nov 2;14(11):2434. doi: 10.3390/v14112434. Viruses. 2022. PMID: 36366532 Free PMC article. Review.
-
Detection and Genomic Characterization of Bovine Rhinitis Virus in China.Animals (Basel). 2023 Jan 16;13(2):312. doi: 10.3390/ani13020312. Animals (Basel). 2023. PMID: 36670851 Free PMC article.
-
Proximity-dependent biotinylation detects associations between SARS coronavirus nonstructural protein 1 and stress granule-associated proteins.J Biol Chem. 2021 Dec;297(6):101399. doi: 10.1016/j.jbc.2021.101399. Epub 2021 Nov 11. J Biol Chem. 2021. PMID: 34774526 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

