miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition
- PMID: 30603114
- PMCID: PMC6308238
- DOI: 10.1038/s41392-018-0033-6
miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition
Abstract
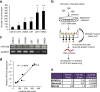
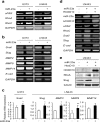
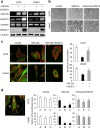
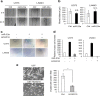
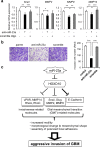
Glioblastoma is the most aggressive and invasive brain tumor and has a poor prognosis; elucidating the underlying molecular mechanisms is essential to select molecular targeted therapies. Here, we investigated the effect of microRNAs on the marked invasiveness of glioblastoma. U373 glioblastoma cells were infected with 140 different microRNAs from an OncomiR library, and the effects of the invasion-related microRNAs and targeted molecules were investigated after repeated Matrigel invasion assays. Screening of the OncomiR library identified miR-23a as a key regulator of glioblastoma invasion. In six glioblastoma cell lines, a positive correlation was detected between the expression levels of miR-23a and invasiveness. A luciferase reporter assay demonstrated that homeobox D10 (HOXD10) was a miR-23a-target molecule, which was verified by high scores from both the PicTar and miRanda algorithms. Forced expression of miR-23a induced expression of invasion-related molecules, including uPAR, RhoA, and RhoC, and altered expression of glial-mesenchymal transition markers such as Snail, Slug, MMP2, MMP9, MMP14, and E-cadherin; however, these changes in expression levels were reversed by HOXD10 overexpression. Thus, miR-23a significantly promoted invasion of glioblastoma cells with polarized formation of focal adhesions, while exogenous HOXD10 overexpression reversed these phenomena. Here, we identify miR-23a-regulated HOXD10 as a pivotal regulator of invasion in glioblastoma, providing a novel mechanism for the aggressive invasiveness of this tumor and providing insight into potential therapeutic targets.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10.Sci Rep. 2013 Dec 5;3:3423. doi: 10.1038/srep03423. Sci Rep. 2013. PMID: 24305689 Free PMC article.
-
Downregulation of microRNA-92b-3p suppresses proliferation, migration, and invasion of gastric cancer SGC-7901 cells by targeting Homeobox D10.J Cell Biochem. 2019 Oct;120(10):17405-17412. doi: 10.1002/jcb.29005. Epub 2019 May 20. J Cell Biochem. 2019. PMID: 31106881
-
microRNA-223 promotes the growth and invasion of glioblastoma cells by targeting tumor suppressor PAX6.Oncol Rep. 2013 Nov;30(5):2263-9. doi: 10.3892/or.2013.2683. Epub 2013 Aug 21. Oncol Rep. 2013. PMID: 23970099
-
Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells.Int J Oncol. 2013 Jul;43(1):63-71. doi: 10.3892/ijo.2013.1935. Epub 2013 May 13. Int J Oncol. 2013. PMID: 23670532
-
miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma.J Gastroenterol Hepatol. 2014 Apr;29(4):835-42. doi: 10.1111/jgh.12429. J Gastroenterol Hepatol. 2014. PMID: 24219032
Cited by
-
Construction of a Novel MYC-Associated ceRNA Regulatory Network to Identify Prognostic Biomarkers in Colon Adenocarcinoma.J Oncol. 2022 Jul 5;2022:3216285. doi: 10.1155/2022/3216285. eCollection 2022. J Oncol. 2022. PMID: 35847359 Free PMC article.
-
The Role of MicroRNAs in Development of Endometrial Cancer: A Literature Review.J Reprod Infertil. 2023 Jul-Sep;24(3):147-165. doi: 10.18502/jri.v24i3.13271. J Reprod Infertil. 2023. PMID: 37663424 Free PMC article. Review.
-
Comprehensive understanding of glioblastoma molecular phenotypes: classification, characteristics, and transition.Cancer Biol Med. 2024 May 6;21(5):363-81. doi: 10.20892/j.issn.2095-3941.2023.0510. Cancer Biol Med. 2024. PMID: 38712813 Free PMC article. Review.
-
Identification of a Novel Metastasis-Related miRNAs-Based Signature for Predicting the Prognosis of Hepatocellular Carcinoma.J Oncol. 2021 Jan 31;2021:6629633. doi: 10.1155/2021/6629633. eCollection 2021. J Oncol. 2021. PMID: 33603784 Free PMC article.
-
miR-5188 augments glioma growth, migration and invasion through an SP1-modulated FOXO1-PI3K/AKT-c-JUN-positive feedback circuit.J Cell Mol Med. 2020 Oct;24(20):11800-11813. doi: 10.1111/jcmm.15794. Epub 2020 Sep 9. J Cell Mol Med. 2020. PMID: 32902145 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

