Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients
- PMID: 30603682
- PMCID: PMC6304452
- DOI: 10.1016/j.heliyon.2018.e01031
Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients
Abstract
Purpose: Although many studies have reported on the resistance mechanism of first-generation EGFR TKIs (1st EGFR TKIs) treatment, large-scale dynamic ctDNA mutation analysis based on liquid biopsy for non-small cell lung cancer (NSCLC) in the Chinese population is rare. Using in-depth integration and analysis of ctDNA genomic mutation data and clinical data at multiple time points during the treatment of 53 NSCLC patients, we described the resistance mechanisms of 1st EGFR TKIs treatment more comprehensively and dynamically. The resulting profile of the polyclonal competitive evolution of the tumor provides some new insights into the precise treatment of NSCLC.
Experimental design: A prospective study was conducted in patients with advanced NSCLC with acquired resistance to erlotinib, gefitinib or icotinib. By liquid biopsy, we detected mutations in 124 tumor-associated genes in the context of drug resistance. These 124 genes covered all tumor therapeutic targets and related biological pathways. During the entire course of treatment, the interval between two liquid biopsies was two months.
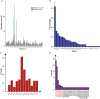
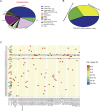
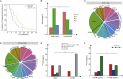
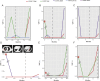
Results: Unlike the common mutations tested in tissue samples, our data showed a higher coverage of tumor heterogeneity (32.65%), more complex patterns of resistance and some new resistance mutation sites, such as EGFR p.V769M and KRAS p.A11V. The major resistance-associated mutations detected were still EGFR p.T790M (45.28%), other point mutations in EGFR (33.9%), and KRAS and NRAS mutations (15.09%). These mutation ratios might be considered as a preliminary summary of the characteristics of Chinese patients. In addition, starting from the two baseline mutations of the EGFR gene (19del vs. L858R), we first described the detailed mutation profile of the EGFR gene. Although there was no significant difference in the number of patients with EGFR p.19del and EGFR p.L858R baseline mutations (24% vs. 16%, P = 0.15), patients from the EGFR p.19del baseline group were much more likely to develop EGFR p.T790M resistance mutations (62.1% vs. 19.3%, P = 0.007). Through careful integration of gene mutation information and clinical phenotype information, an interesting phenomenon was found. Although the variant allele fraction (VAF) of the EGFR p.T790M mutation was significantly linearly correlated with that of the EGFR drug-sensitive mutation (r = 0.68, P = 0.00025), neither VAF was associated with the tumor volume at the advanced stage. It was shown that other tumor clones might contribute more to the resistance to 1st EGFR TKIs treatment than tumor clones carrying the EGFR p.T790M mutation when resistance developed. By further analysis, we found that, in some patients, when the primary tumor clones detected were those carrying EGFR-/- mutations (both types the EGFR p.19del/p.L858R and EGFR p.T790M mutation types were missing), most of them showed a poor prognosis and ineffective late treatment, indicating that EGFR-/- played a more important role than EGFR p.T790M in the process of NSCLC drug resistance in these patients. From the perspective of the clonal evolution of NSCLC tumor cells, these phenomena could be explained by the competitive evolution between different tumor clones. In addition, two new mutations, KRAS p.A11V and EGFR p.V769M, emerged significantly during drug resistance in NSCLC patients and had shown obvious competitive clonal evolution characteristics. Combined with clear clinical drug resistance phenotypic information, we believed that these two new mutations might be related to new drug resistance mechanisms and deserve further study. We have also seen an interesting phenomenon. In some patients undergoing 1st EGFR TKIs treatment, the EGFR p.T790M mutation appeared, disappeared, and reappeared, and this spatial and temporal diversity of the EGFR p.T790M mutation was regulated by targeted drug and chemotherapy and was correlated with the individual tumor mutation profile.
Conclusions: The constitution and competitive evolution of the tumor clones have a decisive influence on treatment and can be regulated by targeted drugs and chemotherapy. Additionally, EGFR p.T790M spatial and temporal diversity during treatment warrants more attention, and this spatial and temporal diversity may be useful for the choice of treatment strategies for certain NSCLC patients. Through longitudinal cfDNA sample analysis, the resistance mechanism and dynamic clinical features of Chinese NSCLC patients are systematically established as reliable and meaningful to understand acquired resistance and make further personalized treatment decisions dynamically. Two new potential drug resistance-associated mutations in EGFR and KRAS have been found and are worthy of further study. Finally, our research shows that the evolutionary process of tumor cloning can be artificially regulated and intervened, possibly providing a new way to treat tumors.
Keywords: Oncology.
Figures




References
-
- Inoue A., Suzuki T., Fukuhara T., Maemondo M., Kimura Y., Morikawa N., Watanabe H., Saijo Y., Nukiwa T. Prospective Phase II study of gefitinib for chemotherapy-naïve patients with advanced non–small-cell lung cancer with epidermal growth factor receptor gene mutations. J. Clin. Oncol. 2006;24:3340–3346. - PubMed
-
- Sequist L.V., Martins R.G., Spigel D., Grunberg S.M., Spira A., Jänne P.A., Joshi V.A., McCollum D., Evans T.L., Muzikansky A., Kuhlmann G.L., Han M., Goldberg J.S. First-line gefitinib in patients with advanced non–small-cell lung cancer harboring somatic EGFR mutations. J. Clin. Oncol. 2008;26:2442–2449. - PubMed
-
- Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E., Palmero R., Garcia-Gomez R., Pallares C., Sanchez J.M. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. - PubMed
-
- Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., Seto T., Satouchi M., Tada H., Hirashima T. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. - PubMed
-
- Lee D.H. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): the road to a success, paved with failures. Pharmacol. Ther. 2017 - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

