A serum-free medium suitable for maintaining cell morphology and liver-specific function in induced human hepatocytes
- PMID: 30603919
- PMCID: PMC6368503
- DOI: 10.1007/s10616-018-0289-2
A serum-free medium suitable for maintaining cell morphology and liver-specific function in induced human hepatocytes
Abstract
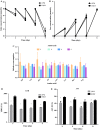
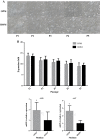
hiHep is a new type of hepatocyte-like cell that is predicted to be a potential unlimited source of hepatocytes for a bioartificial liver. However, hiHep cannot currently be used in clinical settings because serum must be added during the culture process. Thus, a defined medium is required. Because serum is complex, an efficient statistical approach based on the Plackett-Burman design was used. In this manner, an original medium and several significant cell growth factors were identified. These factors include insulin, VH, and VE, and the original medium was optimized based on these significant factors. Additionally, hiHep liver-specific functions and metabolism in the optimized serum-free medium were measured. Results showed that hiHep functions, such as glycogen storage, albumin secretion, and urea production, were well maintained in our optimized serum-free medium. In summary, we created a chemically defined, serum-free medium in which cell growth, proliferation, metabolism, and function were well maintained. This medium has the potential to support the clinical use of hiHep.
Keywords: Bioartificial liver; Medium; Serum-free; hiHep.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources

