A chemical biology toolbox to study protein methyltransferases and epigenetic signaling
- PMID: 30604761
- PMCID: PMC6318333
- DOI: 10.1038/s41467-018-07905-4
A chemical biology toolbox to study protein methyltransferases and epigenetic signaling
Abstract
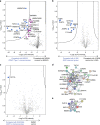
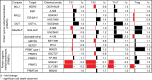
Protein methyltransferases (PMTs) comprise a major class of epigenetic regulatory enzymes with therapeutic relevance. Here we present a collection of chemical probes and associated reagents and data to elucidate the function of human and murine PMTs in cellular studies. Our collection provides inhibitors and antagonists that together modulate most of the key regulatory methylation marks on histones H3 and H4, providing an important resource for modulating cellular epigenomes. We describe a comprehensive and comparative characterization of the probe collection with respect to their potency, selectivity, and mode of inhibition. We demonstrate the utility of this collection in CD4+ T cell differentiation assays revealing the potential of individual probes to alter multiple T cell subpopulations which may have implications for T cell-mediated processes such as inflammation and immuno-oncology. In particular, we demonstrate a role for DOT1L in limiting Th1 cell differentiation and maintaining lineage integrity. This chemical probe collection and associated data form a resource for the study of methylation-mediated signaling in epigenetics, inflammation and beyond.
Conflict of interest statement
Paul L. Richardson is an employee of AbbVie and holds AbbVie stock. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

