Structural and Spectroscopic Characterization of a Product Schiff Base Intermediate in the Reaction of the Quinoprotein Glycine Oxidase, GoxA
- PMID: 30605596
- PMCID: PMC6381839
- DOI: 10.1021/acs.biochem.8b01145
Structural and Spectroscopic Characterization of a Product Schiff Base Intermediate in the Reaction of the Quinoprotein Glycine Oxidase, GoxA
Abstract
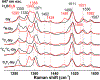
The LodA-like proteins make up a recently identified family of enzymes that rely on a cysteine tryptophylquinone cofactor for catalysis. They differ from other tryptophylquinone enzymes in that they are oxidases rather than dehydrogenases. GoxA is a member of this family that catalyzes the oxidative deamination of glycine. Our previous work with GoxA from Pseudoalteromonas luteoviolacea demonstrated that this protein forms a stable intermediate upon anaerobic incubation with glycine. The spectroscopic properties of this species were unique among those identified for tryptophylquinone enzymes characterized to date. Here we use X-ray crystallography and resonance Raman spectroscopy to identify the GoxA catalytic intermediate as a product Schiff base. Structural work additionally highlights features of the active site pocket that confer substrate specificity, intermediate stabilization, and catalytic activity. The unusual properties of GoxA are discussed within the context of the other tryptophylquinone enzymes.
Figures
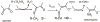








References
-
- McIntire WS, Wemmer DE, Chistoserdov A, and Lidstrom ME (1991) A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase, Science 252, 817–824. - PubMed
-
- Datta S, Mori Y, Takagi K, Kawaguchi K, Chen ZW, Okajima T, Kuroda S, Ikeda T, Kano K, Tanizawa K, and Mathews FS (2001) Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking, Proc. Natl. Acad. Sci. U. S. A 98, 14268–14273. - PMC - PubMed
-
- Satoh A, Kim JK, Miyahara I, Devreese B, Vandenberghe I, Hacisalihoglu A, Okajima T, Kuroda S, Adachi O, Duine JA, Van Beeumen J, Tanizawa K, and Hirotsu K (2002) Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges, J. Biol. Chem 277, 2830–2834. - PubMed
-
- Davidson VL (2005) Structure and mechanism of tryptophylquinone enzymes, Bioorg. Chem 33, 159–170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

