WNT Activates the AAK1 Kinase to Promote Clathrin-Mediated Endocytosis of LRP6 and Establish a Negative Feedback Loop
- PMID: 30605688
- PMCID: PMC6315376
- DOI: 10.1016/j.celrep.2018.12.023
WNT Activates the AAK1 Kinase to Promote Clathrin-Mediated Endocytosis of LRP6 and Establish a Negative Feedback Loop
Abstract
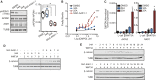
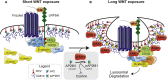
β-Catenin-dependent WNT signal transduction governs development, tissue homeostasis, and a vast array of human diseases. Signal propagation through a WNT-Frizzled/LRP receptor complex requires proteins necessary for clathrin-mediated endocytosis (CME). Paradoxically, CME also negatively regulates WNT signaling through internalization and degradation of the receptor complex. Here, using a gain-of-function screen of the human kinome, we report that the AP2 associated kinase 1 (AAK1), a known CME enhancer, inhibits WNT signaling. Reciprocally, AAK1 genetic silencing or its pharmacological inhibition using a potent and selective inhibitor activates WNT signaling. Mechanistically, we show that AAK1 promotes clearance of LRP6 from the plasma membrane to suppress the WNT pathway. Time-course experiments support a transcription-uncoupled, WNT-driven negative feedback loop; prolonged WNT treatment drives AAK1-dependent phosphorylation of AP2M1, clathrin-coated pit maturation, and endocytosis of LRP6. We propose that, following WNT receptor activation, increased AAK1 function and CME limits WNT signaling longevity.
Keywords: AAK1; AP2M1; LRP6; WNT signaling; clathrin; endocytosis; feedback loop; gain-of-function screen; kinase.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Angers S., Moon R.T. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 2009;10:468–477. - PubMed
-
- Baron C.B., Pompeo J., Blackman D., Coburn R.F. Common phosphatidylinositol 4,5-bisphosphate pools are involved in carbachol and serotonin activation of tracheal smooth muscle. J. Pharmacol. Exp. Ther. 1993;266:8–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 106169/ZZ14/Z/WT_/Wellcome Trust/United Kingdom
- T32 GM007040/GM/NIGMS NIH HHS/United States
- 15291/CRUK_/Cancer Research UK/United Kingdom
- T32 CA071341/CA/NCI NIH HHS/United States
- 24639/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- C7379/A15291/CRUK_/Cancer Research UK/United Kingdom
- U24 DK116204/DK/NIDDK NIH HHS/United States
- R01 CA187799/CA/NCI NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- C7379/A24639/CRUK_/Cancer Research UK/United Kingdom
- T32 CA009156/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R25 GM055336/GM/NIGMS NIH HHS/United States
- F31 CA228289/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

