DBS: a fast and informative segmentation algorithm for DNA copy number analysis
- PMID: 30606105
- PMCID: PMC6318921
- DOI: 10.1186/s12859-018-2565-8
DBS: a fast and informative segmentation algorithm for DNA copy number analysis
Abstract
Background: Genome-wide DNA copy number changes are the hallmark events in the initiation and progression of cancers. Quantitative analysis of somatic copy number alterations (CNAs) has broad applications in cancer research. With the increasing capacity of high-throughput sequencing technologies, fast and efficient segmentation algorithms are required when characterizing high density CNAs data.
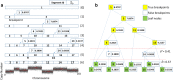
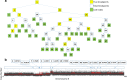
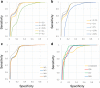
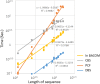
Results: A fast and informative segmentation algorithm, DBS (Deviation Binary Segmentation), is developed and discussed. The DBS method is based on the least absolute error principles and is inspired by the segmentation method rooted in the circular binary segmentation procedure. DBS uses point-by-point model calculation to ensure the accuracy of segmentation and combines a binary search algorithm with heuristics derived from the Central Limit Theorem. The DBS algorithm is very efficient requiring a computational complexity of O(n*log n), and is faster than its predecessors. Moreover, DBS measures the change-point amplitude of mean values of two adjacent segments at a breakpoint, where the significant degree of change-point amplitude is determined by the weighted average deviation at breakpoints. Accordingly, using the constructed binary tree of significant degree, DBS informs whether the results of segmentation are over- or under-segmented.
Conclusion: DBS is implemented in a platform-independent and open-source Java application (ToolSeg), including a graphical user interface and simulation data generation, as well as various segmentation methods in the native Java language.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99(20):12963–12968. doi: 10.1073/pnas.162471999. - DOI - PMC - PubMed
-
- Fridlyand J, Snijders AM, Pinkel D, Albertson DG, Jain AN. Hidden Markov models approach to the analysis of array CGH data. J Multivar Anal. 2004;90(1):132–153. doi: 10.1016/j.jmva.2004.02.008. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

