SIX2 Mediates Late-Stage Metastasis via Direct Regulation of SOX2 and Induction of a Cancer Stem Cell Program
- PMID: 30606720
- PMCID: PMC6586234
- DOI: 10.1158/0008-5472.CAN-18-1791
SIX2 Mediates Late-Stage Metastasis via Direct Regulation of SOX2 and Induction of a Cancer Stem Cell Program
Abstract
The capacity for tumor cells to metastasize efficiently is directly linked to their ability to colonize secondary sites. Here we identify Six2, a developmental transcription factor, as a critical regulator of a breast cancer stem cell program that enables metastatic colonization. In several triple-negative breast cancer (TNBC) models, Six2 enhanced the expression of genes associated with embryonic stem cell programs. Six2 directly bound the Sox2 Srr2 enhancer, promoting Sox2 expression and downstream expression of Nanog, which are both key pluripotency factors. Regulation of Sox2 by Six2 enhanced cancer stem cell properties and increased metastatic colonization. Six2 and Sox2 expression correlated highly in breast cancers including TNBC, where a Six2 expression signature was predictive of metastatic burden and poor clinical outcome. Our findings demonstrate that a SIX2/SOX2 axis is required for efficient metastatic colonization, underscoring a key role for stemness factors in outgrowth at secondary sites. SIGNIFICANCE: These findings provide novel mechanistic insight into stemness and the metastatic outgrowth of triple-negative breast cancer cells.Graphical Abstract: http://cancerres.aacrjournals.org/content/canres/79/4/720/F1.large.jpg.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures
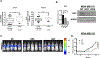
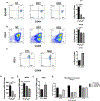

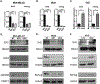



Similar articles
-
MYC Regulates the HIF2α Stemness Pathway via Nanog and Sox2 to Maintain Self-Renewal in Cancer Stem Cells versus Non-Stem Cancer Cells.Cancer Res. 2019 Aug 15;79(16):4015-4025. doi: 10.1158/0008-5472.CAN-18-2847. Epub 2019 Jul 2. Cancer Res. 2019. PMID: 31266772 Free PMC article.
-
Transcription factor Six2 induces a stem cell-like phenotype in renal cell carcinoma cells.FEBS Open Bio. 2019 Oct;9(10):1808-1816. doi: 10.1002/2211-5463.12721. Epub 2019 Sep 19. FEBS Open Bio. 2019. PMID: 31420918 Free PMC article.
-
The FBXW2-MSX2-SOX2 axis regulates stem cell property and drug resistance of cancer cells.Proc Natl Acad Sci U S A. 2019 Oct 8;116(41):20528-20538. doi: 10.1073/pnas.1905973116. Epub 2019 Sep 23. Proc Natl Acad Sci U S A. 2019. PMID: 31548378 Free PMC article.
-
Crosstalks between Raf-kinase inhibitor protein and cancer stem cell transcription factors (Oct4, KLF4, Sox2, Nanog).Tumour Biol. 2017 Apr;39(4):1010428317692253. doi: 10.1177/1010428317692253. Tumour Biol. 2017. PMID: 28378634 Review.
-
SOX2 in development and cancer biology.Semin Cancer Biol. 2020 Dec;67(Pt 1):74-82. doi: 10.1016/j.semcancer.2019.08.007. Epub 2019 Aug 11. Semin Cancer Biol. 2020. PMID: 31412296 Review.
Cited by
-
CAV1 inhibits Xc- system through IFNGR1 to promote ferroptosis to inhibit stemness and improves anti-PD-1 efficacy in breast cancer.Transl Oncol. 2024 Dec;50:102149. doi: 10.1016/j.tranon.2024.102149. Epub 2024 Oct 11. Transl Oncol. 2024. PMID: 39395272 Free PMC article.
-
Targeting SIX2 as a novel sensitization strategy of sorafenib treatment on advanced hepatocellular carcinoma through modulating METTL9-SLC7A11 axis.NPJ Precis Oncol. 2025 Jun 16;9(1):186. doi: 10.1038/s41698-025-01004-6. NPJ Precis Oncol. 2025. PMID: 40523929 Free PMC article.
-
SIX4 Activation in Inflammatory Response Drives the Transformation of Colorectal Epithelium into Inflammation and Tumor via Feedback-Enhancing Inflammatory Signaling to Induce Tumor Stemness Signaling.Int J Biol Sci. 2024 Aug 26;20(12):4618-4634. doi: 10.7150/ijbs.93411. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309424 Free PMC article.
-
Two Sides of the Same Coin: The Role of Developmental pathways and pluripotency factors in normal mammary stem cells and breast cancer metastasis.J Mammary Gland Biol Neoplasia. 2020 Jun;25(2):85-102. doi: 10.1007/s10911-020-09449-0. Epub 2020 Apr 22. J Mammary Gland Biol Neoplasia. 2020. PMID: 32323111 Free PMC article. Review.
-
FZD5 contributes to TNBC proliferation, DNA damage repair and stemness.Cell Death Dis. 2020 Dec 12;11(12):1060. doi: 10.1038/s41419-020-03282-3. Cell Death Dis. 2020. PMID: 33311446 Free PMC article.
References
-
- Chambers AF, Naumov GN, Varghese HJ, Nadkarni KV, MacDonald IC, Groom AC. Critical steps in hematogenous metastasis: an overview. Surg Oncol Clin N Am 2001;10:243–55, vii. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

