Normalization of Gene Expression by Quantitative RT-PCR in Human Cell Line: comparison of 12 Endogenous Reference Genes
- PMID: 30607091
- PMCID: PMC6308757
- DOI: 10.4314/ejhs.v28i6.9
Normalization of Gene Expression by Quantitative RT-PCR in Human Cell Line: comparison of 12 Endogenous Reference Genes
Abstract
Background: Polymerase Chain Reaction (PCR) has become an important diagnostic and research tool of modern molecular biology globally. Real-time PCR allows for rapid and reliable quantification of mRNA transcription. Reference genes are used as internal reaction control to normalise mRNA levels between different samples in order to allow for an exact comparison of mRNA transcription level.
Methods: In this study, twelve commonly used human reference genes were investigated in Human Embryonic Kidney Cell Lines (HEK293) using real-time qPCR with SYBR green. The genes included beta-2-microglobulin (B2M), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), succinate dehydrogenase complex subunit A (SDHA), and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide (YWHAZ). The stability of these reference genes was investigated using the geNorm application.
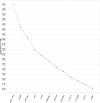
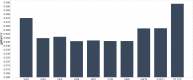
Results: The range of expression stability in the genes analysed was (from the most stable to the least stable): UBC, TOP1, ATP5B, CYC1, GAPDH, SDHA, YWHAZ, CTB, 18S, EIFA-2, B2M and RPL13A. The optimal number of reference targets in the experiment was calculated to be 2 (geNorm V<0.15) when comparing a normalization factor based on the 2 or 3 most stable targets).
Conclusion: The expression stability varied greatly between the 12 candidate reference genes. UBC, TOP1, ATP5B, CYC1 and GAPDH respectively showed the highest stability in HEK293 cells based on both expression stability and expression level. Overall, our data suggest that UBC and TOP1show the least variation and the highest expression stability. This report validates the need for rational selection of reference genes for data normalization to ensure accuracy of quantitative PCR assays.
Keywords: Gene expression; Human cell lines; Nrmalization; PCR; Reference genes.
Figures

Similar articles
-
Pregnancy influences the selection of appropriate reference genes in mouse tissue: Determination of appropriate reference genes for quantitative reverse transcription PCR studies in tissues from the female mouse reproductive axis.Gene. 2021 Oct 30;801:145855. doi: 10.1016/j.gene.2021.145855. Epub 2021 Jul 20. Gene. 2021. PMID: 34293448
-
Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR.BMC Mol Biol. 2007 Aug 15;8:67. doi: 10.1186/1471-2199-8-67. BMC Mol Biol. 2007. PMID: 17697375 Free PMC article.
-
Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue.BMC Mol Biol. 2008 May 6;9:46. doi: 10.1186/1471-2199-9-46. BMC Mol Biol. 2008. PMID: 18460208 Free PMC article.
-
Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model.BMC Mol Biol. 2008 May 28;9:53. doi: 10.1186/1471-2199-9-53. BMC Mol Biol. 2008. PMID: 18505597 Free PMC article.
-
Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines.PLoS One. 2013;8(3):e59180. doi: 10.1371/journal.pone.0059180. Epub 2013 Mar 15. PLoS One. 2013. PMID: 23554992 Free PMC article. Review.
Cited by
-
Selection of suitable reference genes for normalization of RT-qPCR in three tissues of Northern bobwhite (Colinus virginianus) infected with eyeworm (Oxyspirura petrowi).Mol Biol Rep. 2024 Apr 5;51(1):483. doi: 10.1007/s11033-024-09401-z. Mol Biol Rep. 2024. PMID: 38578540
-
Gene expression associated with endocrine therapy resistance in estrogen receptor-positive breast cancer.Sci Rep. 2025 Feb 28;15(1):7220. doi: 10.1038/s41598-025-89274-9. Sci Rep. 2025. PMID: 40021703 Free PMC article.
-
Reference gene validation for the relative quantification of cannabinoid receptor expression in human odontoblasts via quantitative polymerase chain reaction.J Oral Biol Craniofac Res. 2022 Nov-Dec;12(6):765-770. doi: 10.1016/j.jobcr.2022.09.006. Epub 2022 Sep 10. J Oral Biol Craniofac Res. 2022. PMID: 36133217 Free PMC article.
-
Systematic Identification of the Optimal Housekeeping Genes for Accurate Transcriptomic and Proteomic Profiling of Tissues following Complex Traumatic Injury.Methods Protoc. 2023 Feb 23;6(2):22. doi: 10.3390/mps6020022. Methods Protoc. 2023. PMID: 36961042 Free PMC article.
-
A Strategy for the Selection of RT-qPCR Reference Genes Based on Publicly Available Transcriptomic Datasets.Biomedicines. 2023 Apr 3;11(4):1079. doi: 10.3390/biomedicines11041079. Biomedicines. 2023. PMID: 37189697 Free PMC article.
References
-
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. “The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments”. Clinical Chemistry. 2009;55(4):611–622. - PubMed
-
- Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. The real-time polymerase chain reaction. Molecular Aspects of Medicine. 2006;27:95–125. - PubMed
-
- Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of Suitable Reference Genes for qPCR Analysis of Serum microRNA in Gastric Cancer Patients. Digestive Diseases and Sciences. 2012;57(4):897–904. - PubMed
-
- Thellin O, Zorzi W, Lakaye B, DeBorman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. Journal of Biotechnology. 1999;75:291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
